| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,601,896 – 4,602,084 |
| Length | 188 |
| Max. P | 0.996894 |

| Location | 4,601,896 – 4,602,084 |
|---|---|
| Length | 188 |
| Sequences | 3 |
| Columns | 192 |
| Reading direction | forward |
| Mean pairwise identity | 49.73 |
| Shannon entropy | 0.70464 |
| G+C content | 0.48023 |
| Mean single sequence MFE | -55.21 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
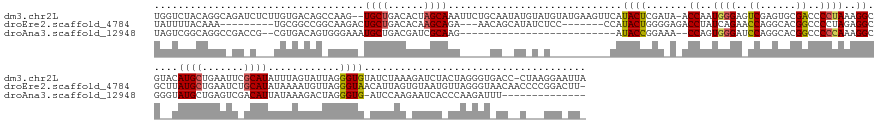
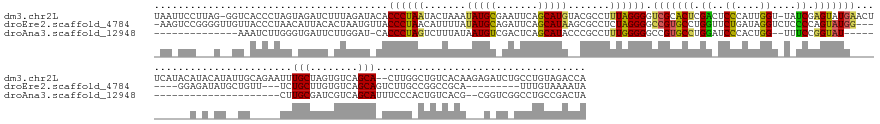
>dm3.chr2L 4601896 188 + 23011544 UGGUCUACAGGCAGAUCUCUUGUGACAGCCAAG--UGCUGACACUAGCAAAUUCUGCAAUAUGUAUGUAUGAAGUUCAUACUCGAUA-ACCAAUGGGAGUCGAGUGCGACCCCUAAAGGCGUACAUGCUGAAUUCGCAUAUUUAGUAUUAGGGUGUAUCUAAAGAUCUACUAGGGUGACC-CUAAGGAAUUA .((((..(.((.((((((......(((.((.((--((((((.....((.(((((.(((...(((((((....((.((.((((((((.-.((...))..)))))))).))...))....)))))))))).))))).))....)))))))).)).)))......)))))).)).)...))))-........... ( -57.20, z-score = -1.83, R) >droEre2.scaffold_4784 23287692 172 + 25762168 UAUUUUACAAA---------UGCGGCCGGCAAGACUGCUGACACAAGCAGA---AACAGCAUAUCUCC-------CCAUACUGGGGAGACCUAUCAGAACCAGGCACGGCCCCUAGAGGCGCUUAUGCUGAAUCUGCAUAUAAAAUGUUAGGGUAACAUUAGUGUAAUGUUAGGGUAACAACCCCGGACUU- ....(((((((---------((.(((((((....(((((......))))).---.........(((((-------((.....)))))))..............)).)))))(((((.......(((((.......))))).......)))))....))))..))))).(((.((((....))))..)))..- ( -53.94, z-score = -1.94, R) >droAna3.scaffold_12948 636040 147 - 692136 UAGUCGGCAGGCCGACCG--CGUGACAGUGGGAAAUGCUGACGAUCGCAAG--------------------------AUACCGGAAA--CCAGUGGGAUCCAGGCACGGCCCCCAAAGGCGGGUAUGCUGAGUCGACAUUAUAAAGACUAGGGUG-AUCCAAGAAUCACCCAAGAUUU-------------- ..((((((.(((((.(((--(......))))....((((....(((....)--------------------------))...(((..--((...))..))).)))))))))(((......)))........))))))..........((.(((((-((......))))))).))....-------------- ( -54.50, z-score = -2.68, R) >consensus UAGUCUACAGGC_GA_C___UGUGACAGCCAAGA_UGCUGACACUAGCAAA_____CA__AU_U_U__________CAUACUGGAAA_ACCAAUGGGAACCAGGCACGGCCCCUAAAGGCGCAUAUGCUGAAUCCGCAUAUUAAAUAUUAGGGUG_AUCUAAAGAUCUACUAGGGUAAC__C___GGA_UU_ .....................((((((((......((((......))))........................................((..((((..((......))..))))..)).......))))...))))....................................................... (-10.80 = -11.70 + 0.90)
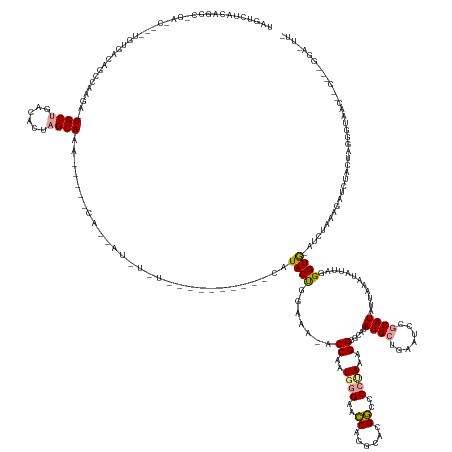
| Location | 4,601,896 – 4,602,084 |
|---|---|
| Length | 188 |
| Sequences | 3 |
| Columns | 192 |
| Reading direction | reverse |
| Mean pairwise identity | 49.73 |
| Shannon entropy | 0.70464 |
| G+C content | 0.48023 |
| Mean single sequence MFE | -57.90 |
| Consensus MFE | -17.96 |
| Energy contribution | -17.97 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
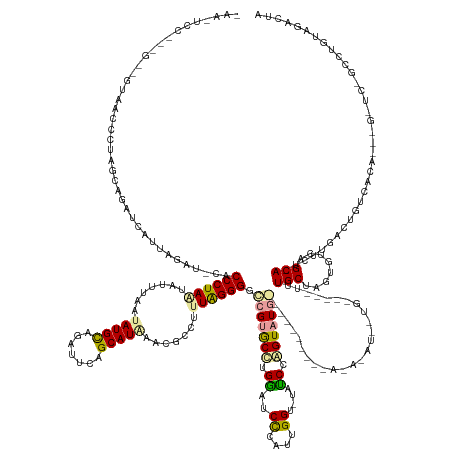
>dm3.chr2L 4601896 188 - 23011544 UAAUUCCUUAG-GGUCACCCUAGUAGAUCUUUAGAUACACCCUAAUACUAAAUAUGCGAAUUCAGCAUGUACGCCUUUAGGGGUCGCACUCGACUCCCAUUGGU-UAUCGAGUAUGAACUUCAUACAUACAUAUUGCAGAAUUUGCUAGUGUCAGCA--CUUGGCUGUCACAAGAGAUCUGCCUGUAGACCA ...........-((((((....((((((((((.......................((((((((.(((((((.(((..(.(((((((....))))))).)..)))-......(((((.....))))).))))...))).))))))))..(((.((((.--....)))).))).))))))))))..)).)))). ( -61.30, z-score = -4.28, R) >droEre2.scaffold_4784 23287692 172 - 25762168 -AAGUCCGGGGUUGUUACCCUAACAUUACACUAAUGUUACCCUAACAUUUUAUAUGCAGAUUCAGCAUAAGCGCCUCUAGGGGCCGUGCCUGGUUCUGAUAGGUCUCCCCAGUAUGG-------GGAGAUAUGCUGUU---UCUGCUUGUGUCAGCAGUCUUGCCGGCCGCA---------UUUGUAAAAUA -......(((((....)))))(((((((...)))))))........((((((((.(((((..(((((((...((((...(((((((....)))))))...))))((((((.....))-------)))).)))))))..---))))).((((((.((......)).)).))))---------..)))))))). ( -61.50, z-score = -2.85, R) >droAna3.scaffold_12948 636040 147 + 692136 --------------AAAUCUUGGGUGAUUCUUGGAU-CACCCUAGUCUUUAUAAUGUCGACUCAGCAUACCCGCCUUUGGGGGCCGUGCCUGGAUCCCACUGG--UUUCCGGUAU--------------------------CUUGCGAUCGUCAGCAUUUCCCACUGUCACG--CGGUCGGCCUGCCGACUA --------------.......((((((((....)))-)))))(((((.......((.((((.(((.(((((.(((..((((..(((....)))..))))..))--)....)))))--------------------------.))).).))).))(((...((.((((.....--)))).))..))).))))) ( -50.90, z-score = -1.75, R) >consensus _AA_UCC___G__GUAACCCUAGCAGAUCAUUAGAU_CACCCUAAUAUUUAAUAUGCAGAUUCAGCAUAAACGCCUUUAGGGGCCGUGCCUGGAUCCCAUUGGU_UAUCCAGUAUG__________A_A_AU__UG_____UUUGCUAGUGUCAGCA_UCUUGACUGUCACA___G_UC_GCCUGUAGACUA .......................................((((((.......(((((.......))))).......)))))).(((((((.((..((....))....)).)))))))..........................(((........)))................................... (-17.96 = -17.97 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:51 2011