
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,186,395 – 8,186,488 |
| Length | 93 |
| Max. P | 0.854357 |

| Location | 8,186,395 – 8,186,488 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 65.22 |
| Shannon entropy | 0.71593 |
| G+C content | 0.43154 |
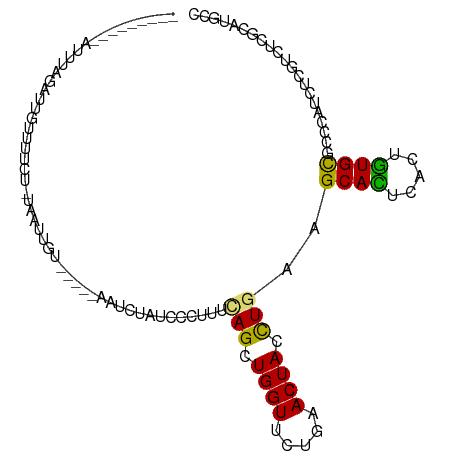
| Mean single sequence MFE | -15.94 |
| Consensus MFE | -8.46 |
| Energy contribution | -8.11 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8186395 93 + 24543557 ---------AUUUAGAUUGUUUUCU-AAAUUGU-----AAUCUAUGCUUUUCAGUUGGUUCUUAACUACCUGAAGCAUUCACUGUGCGCCCAUCUCGUUUCUCACGCU ---------(((((((......)))-)))).((-----(....(((((..((((.((((.....)))).)))))))))......)))..................... ( -16.50, z-score = -1.72, R) >droVir3.scaffold_13049 22603465 104 - 25233164 UUUUAAACAAUUUGCCAUAUCG-CUCUAAUCC---AUUUGUUCUCCUCCUGCAGCUGGUUUUGAACUAUCUGAAGCACUCGCUGUGCGCGCAUCUUGUCUCGCAUGCC ......((((..((((......-.((....((---(.((((.........)))).)))....))..........((((.....))))).)))..)))).......... ( -17.30, z-score = 0.42, R) >droSim1.chr3L 7652273 93 + 22553184 ---------AUUUAGAUUGUUUUCU-AAAUUGU-----AAUUUAUGCCUUUCAGUUGGUUCUUAACUACCUGAAGCACUCACUGUGCGCCCAUCUCGUUUCUCACGCU ---------(((((((......)))-))))...-----.......((..(((((.((((.....)))).)))))((((.....))))..................)). ( -18.00, z-score = -2.24, R) >droSec1.super_0 472452 93 + 21120651 ---------AUUUAGAUUGUUUUCU-AAAUUGU-----AAUUUAUGCCUUUCAGUUGGUUCUUAACUACCUGAAGCACUCACUGUGCGCCCAUCUCGUUUCUCACGCU ---------(((((((......)))-))))...-----.......((..(((((.((((.....)))).)))))((((.....))))..................)). ( -18.00, z-score = -2.24, R) >droYak2.chr3L 20485475 93 - 24197627 ---------AUUUAGUUUGCUUUUA-UAAUAGU-----AAUUUAUUCCGUUCAGUUGGUGCUCAACUACCUAAAGCACUCACUGUGUGCCCAUCUCGUUUCACACGCU ---------...(((.(((((....-....)))-----)).)))........(((.((((((...........)))))).)))(((((............)))))... ( -14.90, z-score = -0.90, R) >droEre2.scaffold_4784 19912740 93 - 25762168 ---------AUUUAGUUUGCUUUCU-UAAUUGU-----AAUUAAUUCUGUUUAGUUGGUGCUGAACUACCUGAAGCACUCACUGUGCGCCCAUCUCGUUUCUCACGCU ---------.(((((.((((.....-.....))-----))........(((((((....)))))))...)))))((((.....))))..................... ( -14.80, z-score = -0.05, R) >droAna3.scaffold_13337 3190907 98 + 23293914 ---AAAUUAAACAAUAAAGAAA--UCUAAAGCA-----AUUCUCUCGUACUCAGCUGGUUCUGAACUACCUGAAGCACUCACUGUGCGCCCAUCUCGUCUCCCAUGCC ---..............(((..--......((.-----............((((.((((.....)))).)))).((((.....))))))........)))........ ( -14.49, z-score = -0.92, R) >dp4.chrXR_group3a 1344100 85 - 1468910 ---------ACUUAAACCAUUAU----GAUUUU-----UGUC-CUCUC----AGCUGGUACUCAACUACCUGAAGCACUCGUUGUGUGCCCACCUCGUCUCGCAUGCC ---------...........(((----(.....-----....-...((----((.((((.....)))).)))).((((.......)))).............)))).. ( -12.00, z-score = 0.19, R) >droPer1.super_23 1534596 85 - 1662726 ---------ACUUAAACCAUUAU----GAUUUU-----UGUC-CUCUC----AGCUGGUACUCAACUACCUGAAGCACUCGUUGUGUGCCCACCUCGUCUCGCAUGCC ---------...........(((----(.....-----....-...((----((.((((.....)))).)))).((((.......)))).............)))).. ( -12.00, z-score = 0.19, R) >droWil1.scaffold_180949 1342678 98 + 6375548 ----AUUUCAUUUUGGCCAUCAUCAGUGAUCAU-----UGUC-AUUUUGUUUAGUUGGUUUUAAACUAUUUGAAGCAUUCGUUAUGCGCUCACCUGGUCUCCCAUGCC ----..........(((((......((((.(..-----..((-(....((((((......))))))....))).((((.....))))).)))).)))))......... ( -16.90, z-score = -0.19, R) >droMoj3.scaffold_6654 1259630 98 + 2564135 ----------AGUGCCAUAGAAUCUCUAAUCCUUCAUUGGCCUCUCUCUUGCAGCUGGUCUUGAACUAUCUGAAGCACUCGCUGUGCGCACAUCUUGUCUCCCAUGCG ----------.((((.........((..((..((((..((((.(((....).))..)))).))))..))..)).((((.....))))))))................. ( -19.40, z-score = 0.17, R) >droGri2.scaffold_15110 20940625 102 - 24565398 ---ACAUCAACCGGAAACCAUUUCUCUAACCCU---UUACUCCUUACCAAGUAGCUGGUGUUGAACUAUCUGAAGCACUCGCUGUGCGCCCAUCUUGUCUCGCAUGCG ---...(((((.(....)..........(((..---(((((........)))))..))))))))...............(((.(((((..(.....)...)))))))) ( -17.00, z-score = -0.02, R) >consensus _________AUUUAGAUUGUUUUCU_UAAUUGU_____AAUCUAUCCCUUUCAGCUGGUUCUGAACUACCUGAAGCACUCACUGUGCGCCCAUCUCGUCUCGCAUGCC ...................................................(((.((((.....)))).)))..((((.....))))..................... ( -8.46 = -8.11 + -0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:11:01 2011