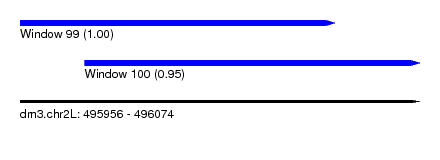
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 495,956 – 496,074 |
| Length | 118 |
| Max. P | 0.998881 |

| Location | 495,956 – 496,049 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Shannon entropy | 0.34351 |
| G+C content | 0.49544 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -23.44 |
| Energy contribution | -25.03 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.998881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
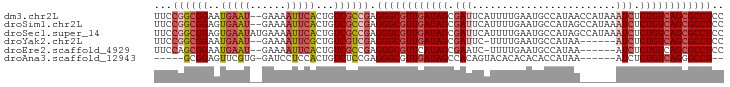
>dm3.chr2L 495956 93 + 23011544 UUCCGGCGGAAUGAAU--GAAAAUUCACUGUCGCCGAGGGCGUUGAUAGCGAUUCAUUUUGAAUGCCAUAACCAUAAAUCUCUGUCAGCGCCUCC ...((((((..(((((--....)))))...)))))).((((((((((((.((((.....((.....))........)))).)))))))))))).. ( -36.22, z-score = -4.67, R) >droSim1.chr2L 502385 93 + 22036055 UUCCGGCGGAGUGAAU--GAAAAUUCACUGUCGCCGAGGGCGUUGAUAGCGAUUCAUUUUGAAUGCCAUAGCCAUAAAUCUCUGUCAGCGCCUCC ...(((((((((((((--....))))))).)))))).((((((((((((.((((.....((...((....))))..)))).)))))))))))).. ( -40.90, z-score = -5.43, R) >droSec1.super_14 478733 95 + 2068291 UUCCGGCGGAGUGAAUAUGAAAAUUCACUGUCGCCGAGGGCGUUGAUAGCGAUUCAUUUUGAAUGCCAUAGCCAUAAAUCUCUGUCAGCGCCUCC ...(((((((((((((......))))))).)))))).((((((((((((.((((.....((...((....))))..)))).)))))))))))).. ( -38.70, z-score = -4.69, R) >droYak2.chr2L 480621 86 + 22324452 UUCCGGCGGAAUGAAU--GAAAAUUCGCUGUCGUCGAGGGCGUUGAUAGCGAUUC-UUUUGAAUGCCAUAA------AUCUCUGUCAGCGCCUCC ...((((((..(((((--....)))))...)))))).((((((((((((.((((.-...((.....))..)------))).)))))))))))).. ( -34.10, z-score = -3.95, R) >droEre2.scaffold_4929 548249 86 + 26641161 UUCCAGCGGAAUGAAU--GAAAAUUCACUGUCGCCGAGGGCGUUCAUAGCGAAUC-UUUUGAAUGCCAUAA------AUCUCUGUCAGCGCCUCC .....((((..(((((--....)))))...))))...(((((((.((((.((...-...((.....))...------.)).)))).))))))).. ( -22.70, z-score = -1.11, R) >droAna3.scaffold_12943 298629 81 - 5039921 -----GCGGAGUUCGUG-GAUCCUCCACUGUCUCCGAGGGCGUUGAUAGCCACAGUACACACACACCAUAA------AUCUCUGUCAGGGCCU-- -----.(((((...(((-(.....))))...))))).((((.(((((((......................------....))))))).))))-- ( -26.47, z-score = -1.53, R) >consensus UUCCGGCGGAAUGAAU__GAAAAUUCACUGUCGCCGAGGGCGUUGAUAGCGAUUCAUUUUGAAUGCCAUAA______AUCUCUGUCAGCGCCUCC ...((((((..(((((......)))))...)))))).((((((((((((.(((........................))).)))))))))))).. (-23.44 = -25.03 + 1.58)
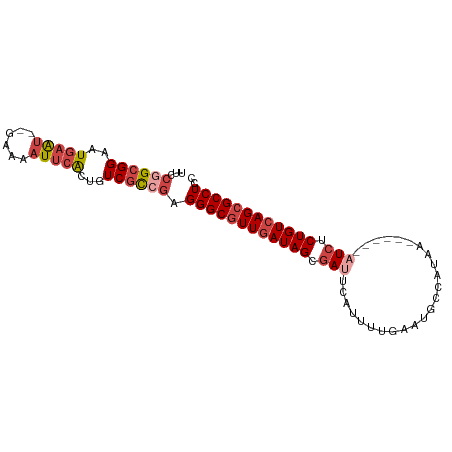
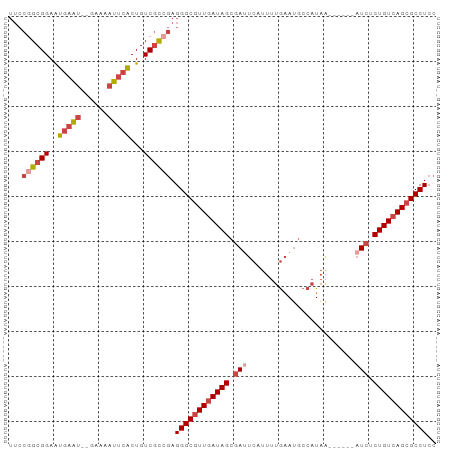
| Location | 495,975 – 496,074 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.20 |
| Shannon entropy | 0.33393 |
| G+C content | 0.49121 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -19.75 |
| Energy contribution | -21.53 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 495975 99 + 23011544 AAUUCACUGUCGCCGAGGGCGUUGAUAGCGAUUCAUUUUGAAUGCCAUAACCAUAAAUCUCUGUCAGCGCCUCCUCGAAACUUCCCAAUCUCGCCGAUA .......(((((.(((((((((((((((.((((.....((.....))........)))).))))))))))).....((....)).....)))).))))) ( -28.12, z-score = -3.28, R) >droSim1.chr2L 502404 99 + 22036055 AAUUCACUGUCGCCGAGGGCGUUGAUAGCGAUUCAUUUUGAAUGCCAUAGCCAUAAAUCUCUGUCAGCGCCUCCUCGAAACUUCCCAAUCUCGCCGAUA .......(((((.(((((((((((((((.((((.....((...((....))))..)))).))))))))))).....((....)).....)))).))))) ( -28.40, z-score = -2.72, R) >droSec1.super_14 478754 99 + 2068291 AAUUCACUGUCGCCGAGGGCGUUGAUAGCGAUUCAUUUUGAAUGCCAUAGCCAUAAAUCUCUGUCAGCGCCUCCUCGAAACUUCCCAAUCUUGCCGAUA .......(((((.(((((((((((((((.((((.....((...((....))))..)))).))))))))))).....((....)).....)))).))))) ( -26.30, z-score = -2.12, R) >droYak2.chr2L 480640 92 + 22324452 AAUUCGCUGUCGUCGAGGGCGUUGAUAGCGAUUC-UUUUGAAUGCCAUA------AAUCUCUGUCAGCGCCUCCUCGAAACUUCCCAAUCUCGCCGAUA .......(((((.(((((((((((((((.((((.-...((.....))..------)))).))))))))))).....((....)).....)))).))))) ( -29.40, z-score = -3.27, R) >droEre2.scaffold_4929 548268 92 + 26641161 AAUUCACUGUCGCCGAGGGCGUUCAUAGCGAAUC-UUUUGAAUGCCAUA------AAUCUCUGUCAGCGCCUCCACGAAACUGCCCAAUCUCGCCGAUA .......(((((.((((((((((((.........-...))))))))...------.......((...((......))..))........)))).))))) ( -20.70, z-score = -0.64, R) >droAna3.scaffold_12943 298644 84 - 5039921 CCUCCACUGUCUCCGAGGGCGUUGAUAGCCACAGUACACACACACCAUA------AAUCUCUGUCAGGGCCUG---GAAACCGACGAAUCUUA------ .......(((((((..((((.(((((((.....................------.....))))))).)))))---))....)))).......------ ( -17.07, z-score = 0.56, R) >consensus AAUUCACUGUCGCCGAGGGCGUUGAUAGCGAUUCAUUUUGAAUGCCAUA______AAUCUCUGUCAGCGCCUCCUCGAAACUUCCCAAUCUCGCCGAUA ........((((.(((((((((((((((.((((......................)))).))))))))))).....(........)...)))).)))). (-19.75 = -21.53 + 1.78)

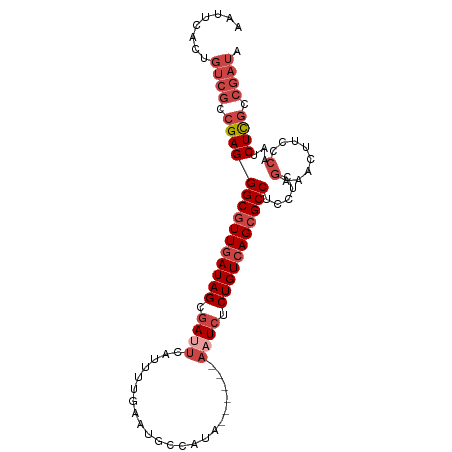
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:02 2011