
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,059,093 – 8,059,198 |
| Length | 105 |
| Max. P | 0.779347 |

| Location | 8,059,093 – 8,059,198 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.89 |
| Shannon entropy | 0.24132 |
| G+C content | 0.55265 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -25.66 |
| Energy contribution | -25.52 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8059093 105 + 24543557 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACACAUGGGAUGAG-CAUACCAAAU ((((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))))..(((......-....)))... ( -29.70, z-score = -1.51, R) >droSim1.chr3L 7522869 105 + 22553184 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACACAUGGGAUGAG-CAUACCAAAU ((((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))))..(((......-....)))... ( -29.70, z-score = -1.51, R) >droSec1.super_0 345795 105 + 21120651 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACACAUGGGAUGAG-UAUACAAAAU ((((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))))...........-.......... ( -27.80, z-score = -1.07, R) >droYak2.chr3L 8645671 105 + 24197627 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACACAUGAGAUGAG-CAUACCAAAU ((((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))))...........-.......... ( -27.80, z-score = -1.76, R) >droEre2.scaffold_4784 22456331 105 - 25762168 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACACAUAAGAUGAG-CAUACCAAAU ((((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))))...........-.......... ( -27.80, z-score = -2.12, R) >droAna3.scaffold_13337 9703669 105 - 23293914 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACAUCUGGGGUGAG-CGCACCAAAU ((((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....)))))).....((((..-..)))).... ( -32.90, z-score = -1.02, R) >dp4.chrXR_group8 5960775 106 + 9212921 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAAGUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUGGGAUGAG-CAGACCAAAA .(((....(((((.......)))))((((((....)))).(((((.......))))).))....)))...((.(((((((..(...)..))))).)-).))...... ( -31.90, z-score = -1.45, R) >droPer1.super_45 100258 106 - 618639 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAAGUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUGGGAUGAG-CAGACCAAAA .(((....(((((.......)))))((((((....)))).(((((.......))))).))....)))...((.(((((((..(...)..))))).)-).))...... ( -31.90, z-score = -1.45, R) >droWil1.scaffold_180727 15928 105 + 2741493 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUA-CAGCGGCCGAGCCAAAU .(((....(((((.......))))).......((-(..(.(((((.......))))))..))).)))..(((((((...(((.......-))).)))))))...... ( -33.70, z-score = -2.03, R) >droVir3.scaffold_13049 12997701 105 - 25233164 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACAGCUGGCAUGAGUCGAGCCAAU- ((((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))))((((((....))).)))....- ( -32.80, z-score = -1.25, R) >droMoj3.scaffold_6680 8608214 105 + 24764193 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUGGCAUGAGUUGAGCCAAA- .(((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....)))))..(((((.........))))).- ( -32.20, z-score = -1.29, R) >droGri2.scaffold_15110 12917479 105 - 24565398 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUGGCAUGAGGUGCGCCAGC- ..((((..(((((.......)))))((((((...-))))...))...))))..((.((((((((..(((..(((.(.....).))).)))..))))))))))....- ( -34.10, z-score = -0.69, R) >anoGam1.chr2R 37004334 83 - 62725911 UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUACGACCUGAACGUAGCGCCUUAGACCGCUCGGCCAUCCUGAC----------------------- .(((((..(((((.......)))))(((..(((.-((........)))))...(.((((........))))))))....)))))----------------------- ( -24.40, z-score = -1.52, R) >consensus UGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA_GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACAGAUGGGAUGAG_CAUACCAAAU .(((((..(((((.......)))))((((((....)))(.(((((.......))))))..............)))....)))))....................... (-25.66 = -25.52 + -0.14)


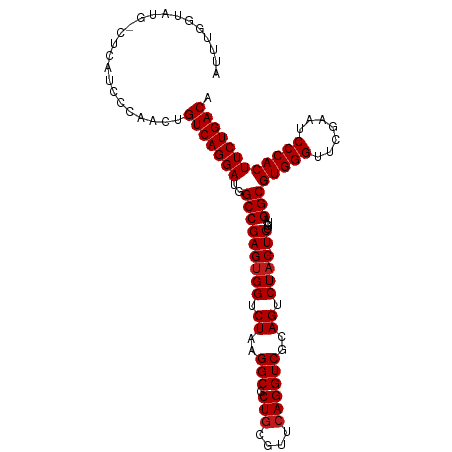
| Location | 8,059,093 – 8,059,198 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.89 |
| Shannon entropy | 0.24132 |
| G+C content | 0.55265 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -32.54 |
| Energy contribution | -32.40 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.779347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8059093 105 - 24543557 AUUUGGUAUG-CUCAUCCCAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA ...(((.((.-...)).)))..((((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......))))))))))))) ( -37.30, z-score = -1.27, R) >droSim1.chr3L 7522869 105 - 22553184 AUUUGGUAUG-CUCAUCCCAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA ...(((.((.-...)).)))..((((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......))))))))))))) ( -37.30, z-score = -1.27, R) >droSec1.super_0 345795 105 - 21120651 AUUUUGUAUA-CUCAUCCCAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA ..........-...........((((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......))))))))))))) ( -36.00, z-score = -1.56, R) >droYak2.chr3L 8645671 105 - 24197627 AUUUGGUAUG-CUCAUCUCAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA ..........-...........((((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......))))))))))))) ( -36.00, z-score = -1.13, R) >droEre2.scaffold_4784 22456331 105 + 25762168 AUUUGGUAUG-CUCAUCUUAUGUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA ..........-...........((((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......))))))))))))) ( -36.00, z-score = -1.34, R) >droAna3.scaffold_13337 9703669 105 + 23293914 AUUUGGUGCG-CUCACCCCAGAUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA ((((((.(..-....).))))))(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))). ( -39.70, z-score = -1.15, R) >dp4.chrXR_group8 5960775 106 - 9212921 UUUUGGUCUG-CUCAUCCCAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA ..((((..(.-...)..))))..(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......)))))))))))). ( -35.80, z-score = -0.91, R) >droPer1.super_45 100258 106 + 618639 UUUUGGUCUG-CUCAUCCCAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA ..((((..(.-...)..))))..(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......)))))))))))). ( -35.80, z-score = -0.91, R) >droWil1.scaffold_180727 15928 105 - 2741493 AUUUGGCUCGGCCGCUG-UAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA .....((((((((((((-.......)))..))))))))).(((....((((((((.......))))))((((-...))))))(((((.......)))))....))). ( -40.30, z-score = -1.36, R) >droVir3.scaffold_13049 12997701 105 + 25233164 -AUUGGCUCGACUCAUGCCAGCUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA -.(((((.........)))))..(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))). ( -41.20, z-score = -1.44, R) >droMoj3.scaffold_6680 8608214 105 - 24764193 -UUUGGCUCAACUCAUGCCAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA -.(((((.........)))))..(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))). ( -41.60, z-score = -2.48, R) >droGri2.scaffold_15110 12917479 105 + 24565398 -GCUGGCGCACCUCAUGCCAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA -(.(((((.......))))).).(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))). ( -41.10, z-score = -1.29, R) >anoGam1.chr2R 37004334 83 + 62725911 -----------------------GUCAGGAUGGCCGAGCGGUCUAAGGCGCUACGUUCAGGUCGUAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA -----------------------(((((((((((((((((.............))))).)))))).((((((-...))))))(((((.......))))).)))))). ( -34.12, z-score = -2.58, R) >consensus AUUUGGUAUG_CUCAUCCCAACUGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC_UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACA .......................(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......)))))))))))). (-32.54 = -32.40 + -0.14)


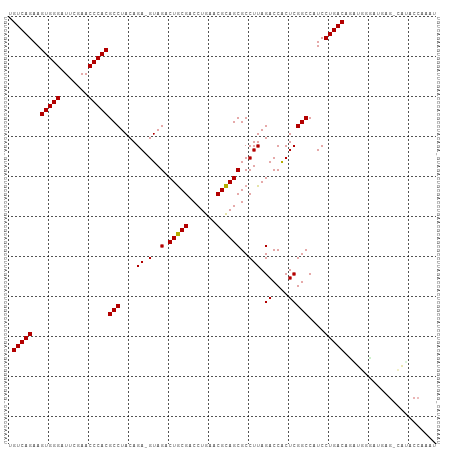
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:52 2011