
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,058,719 – 8,058,792 |
| Length | 73 |
| Max. P | 0.995714 |

| Location | 8,058,719 – 8,058,792 |
|---|---|
| Length | 73 |
| Sequences | 11 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Shannon entropy | 0.42184 |
| G+C content | 0.44177 |
| Mean single sequence MFE | -17.13 |
| Consensus MFE | -12.66 |
| Energy contribution | -12.76 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
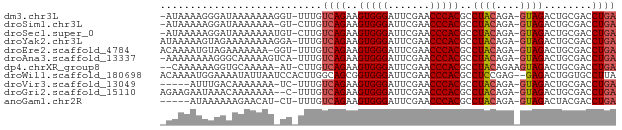
>dm3.chr3L 8058719 73 + 24543557 -AUAAAAGGGAUAAAAAAAGGU-UUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGA -.....((((((((((.....)-))))))....(((((.......))))).))).(((.-((........))))). ( -17.20, z-score = -1.17, R) >droSim1.chr3L 7522514 72 + 22553184 -AUAAAAAGGAUAAAAAAA-GU-CUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGA -......(((........(-((-(((((.((..(((((.......)))))..)))))..-..)))))....))).. ( -15.10, z-score = -1.10, R) >droSec1.super_0 345464 73 + 21120651 -AUAAAAAGGAUAAAAAAAUGU-CUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGA -......((((((......)))-))).((((..(((((.......)))))((((((...-))))...))...)))) ( -16.10, z-score = -1.47, R) >droYak2.chr3L 8645307 74 + 24197627 AUAAAAAGUAGAAAAAAAAGGA-UUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGA ..................(((.-.(((.(((..(((((.......)))))..((((...-)))).))))))))).. ( -15.40, z-score = -1.53, R) >droEre2.scaffold_4784 22455991 73 - 25762168 ACAAAAUGUAGAAAAAAA-GGU-UUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGA .................(-(((-(..(.(((..(((((.......)))))..((((...-)))).))))))))).. ( -16.20, z-score = -1.06, R) >droAna3.scaffold_13337 9703184 73 - 23293914 -AAAAAAAAGGGCAAAAAGUCA-UUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGA -.......(((......((((.-(((((.((..(((((.......)))))..)))))))-...))))....))).. ( -19.50, z-score = -1.88, R) >dp4.chrXR_group8 5954112 72 + 9212921 --CAAAAAAGGUGCAAAAA-AU-CUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAAGUAGACUGCGACCUGA --......(((((((....-..-..........(((((.......)))))..((((....))))..))).)))).. ( -18.60, z-score = -2.03, R) >droWil1.scaffold_180698 4703369 74 - 11422946 ACAAAAUGGAAAAUAUUAAUCCACUUGGCAGCGGUGGGAUUCGAACCCACGCCUCCGAG--GAGACUGGUGCCUUA ......((((.........))))...((((.(((((((.......)))....(((....--))))))).))))... ( -21.40, z-score = -1.13, R) >droVir3.scaffold_13049 12997683 68 - 25233164 -----AUUUGACAAAAAAA-UC-UUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGA -----.(((((((((....-..-))))))))).(((((.......))))).....(((.-((........))))). ( -18.70, z-score = -2.64, R) >droGri2.scaffold_15110 12917457 72 - 24565398 AGAAGAAUAAACAAAAAAA--C-UUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGA ..................(--(-(((((.((..(((((.......)))))..)))))))-)).............. ( -16.00, z-score = -2.12, R) >anoGam1.chr2R 37003819 68 - 62725911 -----AUAAAAAAGAACAU-CU-UUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUACGACCUGA -----....(((((.....-))-))).((((..(((((.......)))))..((((...-))))........)))) ( -14.20, z-score = -1.32, R) >consensus _AAAAAAAAGAAAAAAAAA_GU_UUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA_GUAGACUGCGACCUGA ...........................((((..(((((.......)))))..((((....))))........)))) (-12.66 = -12.76 + 0.11)
| Location | 8,058,719 – 8,058,792 |
|---|---|
| Length | 73 |
| Sequences | 11 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Shannon entropy | 0.42184 |
| G+C content | 0.44177 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
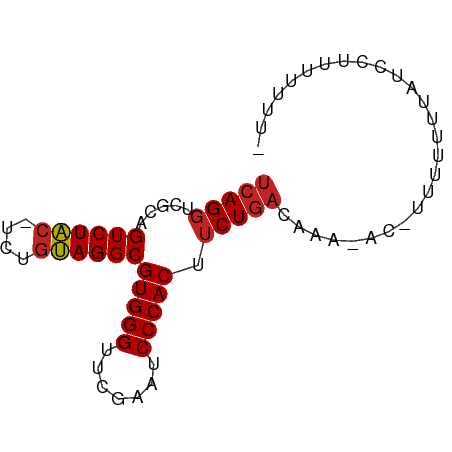
>dm3.chr3L 8058719 73 - 24543557 UCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAA-ACCUUUUUUUAUCCCUUUUAU- (((((.....((((((-...))))))(((((.......))))).)))))....-.....................- ( -17.60, z-score = -2.08, R) >droSim1.chr3L 7522514 72 - 22553184 UCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAG-AC-UUUUUUUAUCCUUUUUAU- ..(((((...((((((-...))))))(((((.......))))).........)-))-))................- ( -18.30, z-score = -2.08, R) >droSec1.super_0 345464 73 - 21120651 UCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAG-ACAUUUUUUUAUCCUUUUUAU- (((((.....((((((-...))))))(((((.......))))).)))))....-.....................- ( -17.60, z-score = -1.89, R) >droYak2.chr3L 8645307 74 - 24197627 UCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAA-UCCUUUUUUUUCUACUUUUUAU (((((.....((((((-...))))))(((((.......))))).)))))....-...................... ( -17.60, z-score = -2.52, R) >droEre2.scaffold_4784 22455991 73 + 25762168 UCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAA-ACC-UUUUUUUCUACAUUUUGU (((((.....((((((-...))))))(((((.......))))).)))))....-...-.................. ( -17.60, z-score = -1.78, R) >droAna3.scaffold_13337 9703184 73 + 23293914 UCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAA-UGACUUUUUGCCCUUUUUUUU- (((.(((...((((((-...))))))(((((.......)))))....)))...-)))..................- ( -18.80, z-score = -1.63, R) >dp4.chrXR_group8 5954112 72 - 9212921 UCAGGUCGCAGUCUACUUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAG-AU-UUUUUGCACCUUUUUUG-- ..((((.(((((((((....))))))(((((.......))))).(((....))-).-....)))))))......-- ( -23.30, z-score = -3.49, R) >droWil1.scaffold_180698 4703369 74 + 11422946 UAAGGCACCAGUCUC--CUCGGAGGCGUGGGUUCGAAUCCCACCGCUGCCAAGUGGAUUAAUAUUUUCCAUUUUGU ...((((.(.(((((--....)))))(((((.......))))).).))))(((((((.........)))))))... ( -27.60, z-score = -3.31, R) >droVir3.scaffold_13049 12997683 68 + 25233164 UCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAA-GA-UUUUUUUGUCAAAU----- ....(((((((.....-.)))).)))(((((.......)))))...(((((((-(.-...))))))))...----- ( -20.40, z-score = -2.72, R) >droGri2.scaffold_15110 12917457 72 + 24565398 UCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAA-G--UUUUUUUGUUUAUUCUUCU (((((.....((((((-...))))))(((((.......))))).)))))....-.--................... ( -17.60, z-score = -1.96, R) >anoGam1.chr2R 37003819 68 + 62725911 UCAGGUCGUAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAA-AG-AUGUUCUUUUUUAU----- (((((.....((((((-...))))))(((((.......))))).))))).(((-((-.....)))))....----- ( -18.80, z-score = -2.24, R) >consensus UCAGGUCGCAGUCUAC_UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAA_AC_UUUUUUUAUCCUUUUUUU_ (((((.....((((((....))))))(((((.......))))).)))))........................... (-16.88 = -17.08 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:50 2011