
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,053,539 – 8,053,638 |
| Length | 99 |
| Max. P | 0.986228 |

| Location | 8,053,539 – 8,053,638 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Shannon entropy | 0.38653 |
| G+C content | 0.50348 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -20.01 |
| Energy contribution | -20.75 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
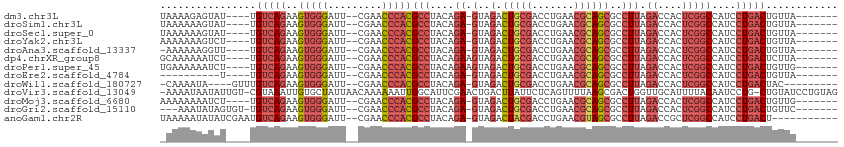
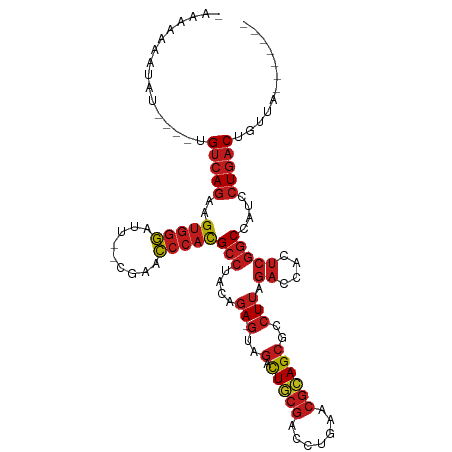
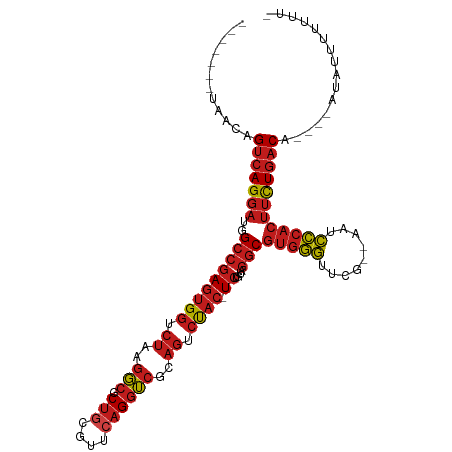
>dm3.chr3L 8053539 99 + 24543557 UAAAAGAGUAU----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUA------- ...........----.(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.90, z-score = -1.33, R) >droSim1.chr3L 7517486 99 + 22553184 UAAAAAAGUAU----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUA------- ...........----.(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.90, z-score = -1.66, R) >droSec1.super_0 340483 99 + 21120651 UAAAAAAGUAU----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUA------- ...........----.(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.90, z-score = -1.66, R) >droYak2.chr3L 8640176 99 + 24197627 AAAAAAAGUCU----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUA------- ...........----.(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.90, z-score = -1.25, R) >droAna3.scaffold_13337 9698173 98 - 23293914 -AAAAAAGGUU----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUA------- -..........----.(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.90, z-score = -0.93, R) >dp4.chrXR_group8 5954122 100 + 9212921 GCAAAAAAUCU----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGAAGUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUCUUA------- ...........----.(((((..(((((...--....)))))(((.....(((..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.60, z-score = -1.61, R) >droPer1.super_45 94685 100 - 618639 UGAAAAAAUCU----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGAAGUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUG------- ...........----.(((((..(((((...--....)))))(((.....(((..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.60, z-score = -1.55, R) >droEre2.scaffold_4784 22450930 89 - 25762168 ----------U----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUA------- ----------.----.(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.90, z-score = -1.78, R) >droWil1.scaffold_180727 6979 96 + 2741493 -CAAAAUA----GUUUGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUAC--------- -.......----....(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....)))))...--------- ( -26.90, z-score = -1.93, R) >droVir3.scaffold_13049 20892980 110 + 25233164 -AAAAUAAUAUUGU-CGUAAAUUGUGCUAUUAACAAAAAAUUGGCAUUCGAACUGACUUAUUCUCAGUUUUAAGCGACUGGUUGCAUUUUACAAUCCUG-CUGUAUCCUGUAG -...........((-(((.....((((((............))))))..(((((((.......)))))))...))))).(((((.......)))))(((-(........)))) ( -20.20, z-score = -1.21, R) >droMoj3.scaffold_6680 8608203 99 + 24764193 AAAAAAAAUCU----UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUG------- ...........----.(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.90, z-score = -1.82, R) >droGri2.scaffold_15110 12912649 99 - 24565398 ---AAAUAUAGUGU-UGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUC------- ---...........-.(((((..(((((...--....)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).....------- ( -26.90, z-score = -0.97, R) >anoGam1.chr2R 37003154 99 - 62725911 UAAAAAUAUAUCGAAUGUCAGAAGUGGGAUU--CGAACCCACGCCUACAGA-GUAGACUACGACCUGAACGUAGCGCCUUAGACCGCUCGGCCAUCCUGACU----------- ................(((((..(((((...--....)))))(((..(((.-((........)))))...(.((((........))))))))....))))).----------- ( -25.60, z-score = -1.85, R) >consensus _AAAAAAAUAU____UGUCAGAAGUGGGAUU__CGAACCCACGCCUACAGA_GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUA_______ ................(((((..(((((.........)))))((((((....)))(.(((((.......))))))..............)))....)))))............ (-20.01 = -20.75 + 0.74)
| Location | 8,053,539 – 8,053,638 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Shannon entropy | 0.38653 |
| G+C content | 0.50348 |
| Mean single sequence MFE | -34.01 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.81 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
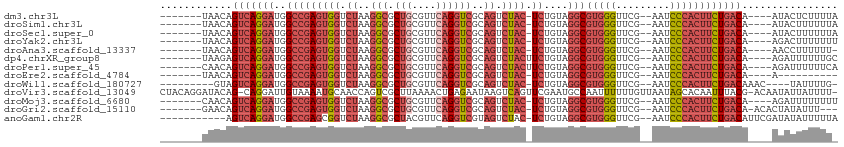
>dm3.chr3L 8053539 99 - 24543557 -------UAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----AUACUCUUUUA -------.....(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...)))))))))))).----........... ( -36.10, z-score = -2.48, R) >droSim1.chr3L 7517486 99 - 22553184 -------UAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----AUACUUUUUUA -------.....(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...)))))))))))).----........... ( -36.10, z-score = -2.70, R) >droSec1.super_0 340483 99 - 21120651 -------UAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----AUACUUUUUUA -------.....(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...)))))))))))).----........... ( -36.10, z-score = -2.70, R) >droYak2.chr3L 8640176 99 - 24197627 -------UAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----AGACUUUUUUU -------.....(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...)))))))))))).----........... ( -36.10, z-score = -2.28, R) >droAna3.scaffold_13337 9698173 98 + 23293914 -------UAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----AACCUUUUUU- -------.....(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...)))))))))))).----..........- ( -36.10, z-score = -2.53, R) >dp4.chrXR_group8 5954122 100 - 9212921 -------UAAGAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUUCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----AGAUUUUUUGC -------.....(((((((.((((....))))....((((((((.......))))))((((....))))))(((((....--...)))))))))))).----........... ( -34.30, z-score = -1.45, R) >droPer1.super_45 94685 100 + 618639 -------CAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUUCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----AGAUUUUUUCA -------.....(((((((.((((....))))....((((((((.......))))))((((....))))))(((((....--...)))))))))))).----........... ( -34.30, z-score = -1.80, R) >droEre2.scaffold_4784 22450930 89 + 25762168 -------UAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----A---------- -------.....(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...)))))))))))).----.---------- ( -36.10, z-score = -2.79, R) >droWil1.scaffold_180727 6979 96 - 2741493 ---------GUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACAAAC----UAUUUUG- ---------...(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...))))))))))))....----.......- ( -36.10, z-score = -2.63, R) >droVir3.scaffold_13049 20892980 110 - 25233164 CUACAGGAUACAG-CAGGAUUGUAAAAUGCAACCAGUCGCUUAAAACUGAGAAUAAGUCAGUUCGAAUGCCAAUUUUUUGUUAAUAGCACAAUUUACG-ACAAUAUUAUUUU- ....((((((...-..((.((((.....)))))).(((((((..((((((.......))))))..)).))........(((.....)))........)-)).....))))))- ( -14.30, z-score = 0.87, R) >droMoj3.scaffold_6680 8608203 99 - 24764193 -------CAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA----AGAUUUUUUUU -------.....(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...)))))))))))).----........... ( -36.10, z-score = -2.58, R) >droGri2.scaffold_15110 12912649 99 + 24565398 -------GAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACA-ACACUAUAUUU--- -------.....(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((....--...)))))))))))).-...........--- ( -36.10, z-score = -2.28, R) >anoGam1.chr2R 37003154 99 + 62725911 -----------AGUCAGGAUGGCCGAGCGGUCUAAGGCGCUACGUUCAGGUCGUAGUCUAC-UCUGUAGGCGUGGGUUCG--AAUCCCACUUCUGACAUUCGAUAUAUUUUUA -----------.(((((((((((((((((.............))))).)))))).((((((-...))))))(((((....--...))))).))))))................ ( -34.32, z-score = -2.30, R) >consensus _______UAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC_UCUGUAGGCGUGGGUUCG__AAUCCCACUUCUGACA____AUAUUUUUUU_ ............(((((((.......(((..((((.((...)).)).))..))).((((((....))))))(((((.........))))))))))))................ (-26.47 = -26.81 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:48 2011