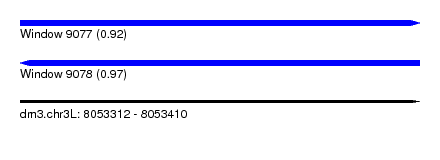
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,053,312 – 8,053,410 |
| Length | 98 |
| Max. P | 0.972223 |

| Location | 8,053,312 – 8,053,410 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 89.45 |
| Shannon entropy | 0.25348 |
| G+C content | 0.54387 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -25.43 |
| Energy contribution | -25.25 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
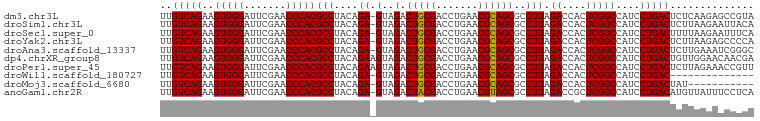
>dm3.chr3L 8053312 98 + 24543557 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUCUCAAGAGCCGUA ..(((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).............. ( -26.90, z-score = -0.59, R) >droSim1.chr3L 7517247 98 + 22553184 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUCUUAAGAAUUACA ..(((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).............. ( -26.90, z-score = -1.81, R) >droSec1.super_0 340241 98 + 21120651 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUUUUAAGAAUUUCA ..(((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).............. ( -26.90, z-score = -1.81, R) >droYak2.chr3L 8639933 98 + 24197627 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUCUUAAGAGCCCCA ..(((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).............. ( -26.90, z-score = -1.25, R) >droAna3.scaffold_13337 9697953 98 - 23293914 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUCUUGAAAUCGGGC ..(((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....))))).((((....)))). ( -27.90, z-score = -0.61, R) >dp4.chrXR_group8 5953855 99 + 9212921 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAAGUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUGUUGGAACAACGA ..(((((..(((((.......)))))(((.....(((..(.(((((.......))))))..))).((....)))))....)))))(((((...))))). ( -27.90, z-score = -1.17, R) >droPer1.super_45 93211 99 - 618639 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAAGUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUCUUAGAAACCGUU ..(((((..(((((.......)))))(((.....(((..(.(((((.......))))))..))).((....)))))....))))).............. ( -26.60, z-score = -1.28, R) >droWil1.scaffold_180727 6135 84 + 2741493 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGAC-------------- ..(((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....)))))-------------- ( -25.70, z-score = -1.96, R) >droMoj3.scaffold_6680 8603603 87 + 24764193 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUAU----------- ..(((((..(((((.......)))))(((....((-(..(.(((((.......))))))..))).((....)))))....)))))...----------- ( -26.90, z-score = -2.39, R) >anoGam1.chr2R 37002203 98 - 62725911 UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA-GUAGACUACGACCUGAACGUAGCGCCUUAGACCGCUCGGCCAUCCUGACAUGUUAUUUCCUCA .((((((..(((((.......)))))(((..(((.-((........)))))...(.((((........))))))))....))))))............. ( -26.80, z-score = -1.46, R) >consensus UUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGA_GUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUCUUAAGAACCGCA ..(((((..(((((.......)))))((((((....)))(.(((((.......))))))..............)))....))))).............. (-25.43 = -25.25 + -0.18)
| Location | 8,053,312 – 8,053,410 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.45 |
| Shannon entropy | 0.25348 |
| G+C content | 0.54387 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -32.73 |
| Energy contribution | -32.55 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
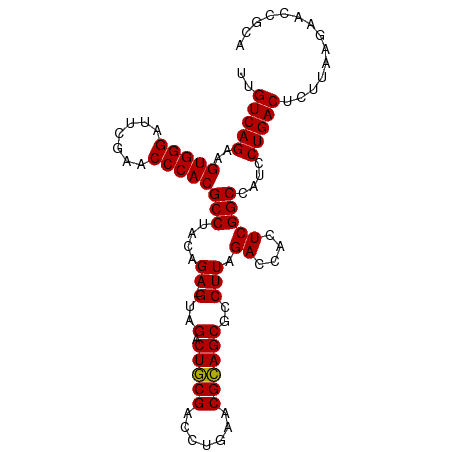
>dm3.chr3L 8053312 98 - 24543557 UACGGCUCUUGAGAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA ..............(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))).. ( -36.10, z-score = -0.99, R) >droSim1.chr3L 7517247 98 - 22553184 UGUAAUUCUUAAGAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA ..............(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))).. ( -36.10, z-score = -2.33, R) >droSec1.super_0 340241 98 - 21120651 UGAAAUUCUUAAAAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA ..............(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))).. ( -36.10, z-score = -2.69, R) >droYak2.chr3L 8639933 98 - 24197627 UGGGGCUCUUAAGAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA ..............(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))).. ( -36.10, z-score = -1.09, R) >droAna3.scaffold_13337 9697953 98 + 23293914 GCCCGAUUUCAAGAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA ..............(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))).. ( -36.10, z-score = -1.45, R) >dp4.chrXR_group8 5953855 99 - 9212921 UCGUUGUUCCAACAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA ..((((...)))).(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......)))))))))))).. ( -35.80, z-score = -1.84, R) >droPer1.super_45 93211 99 + 618639 AACGGUUUCUAAGAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA ..............(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......)))))))))))).. ( -34.30, z-score = -1.25, R) >droWil1.scaffold_180727 6135 84 - 2741493 --------------GUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA --------------(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))).. ( -35.90, z-score = -2.92, R) >droMoj3.scaffold_6680 8603603 87 - 24764193 -----------AUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA -----------...(((((((..(((((((((.((..(((.(((....))))))..)).))))-))....)))(((((.......)))))))))))).. ( -36.10, z-score = -2.93, R) >anoGam1.chr2R 37002203 98 + 62725911 UGAGGAAAUAACAUGUCAGGAUGGCCGAGCGGUCUAAGGCGCUACGUUCAGGUCGUAGUCUAC-UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA .............((((((((((((((((((.............))))).)))))).((((((-...))))))(((((.......))))).))))))). ( -35.12, z-score = -2.01, R) >consensus UGCGGUUCUUAAGAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUAC_UCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAA ..............(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......)))))))))))).. (-32.73 = -32.55 + -0.18)

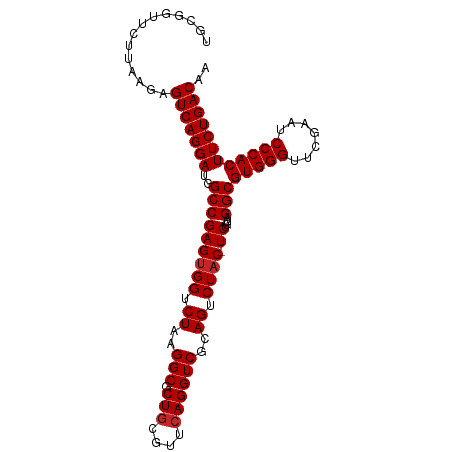
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:46 2011