
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,029,873 – 8,030,003 |
| Length | 130 |
| Max. P | 0.681420 |

| Location | 8,029,873 – 8,030,003 |
|---|---|
| Length | 130 |
| Sequences | 5 |
| Columns | 134 |
| Reading direction | forward |
| Mean pairwise identity | 83.91 |
| Shannon entropy | 0.27400 |
| G+C content | 0.43319 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -21.35 |
| Energy contribution | -21.19 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
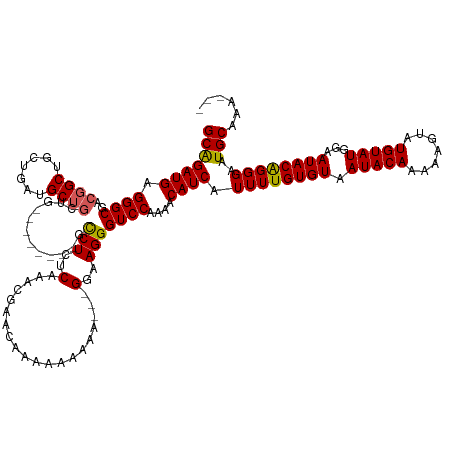
>dm3.chr3L 8029873 130 + 24543557 GCAGAUGAGGGCGACGGCUGCUGAUGCUGCUGGCUGGCUGGCCUCUCAAACGAAAAAAAAAAGA---GGAAGGGUCCAAAACAUCAUUUUGUGUAAUACAAAAAGUAUGUAUGGAAUACAGGGAAUGCAAAAA- (((((((.((((..((((.((....)).)))).....((..(((((...............)))---))..))))))....)))).((((((((.(((((.......)))))...))))))))..))).....- ( -33.96, z-score = -1.45, R) >droSim1.chr3L 7493826 118 + 22553184 GCGGAUGAGGGCGACGGCUGCUGAUGCUGCUG--------GCCUCUCAAACGAACAAAAAAAAAA--GGAAGGGUCCAAAACAUCAUUUUGUGUAAUACAAAAAGUAUGUAUGGAAUACAGGGAAUGC------ .((..(((((((..((((.((....)).))))--------))).))))..)).............--(((....))).......((((((.(((((((((.......)))))....)))).)))))).------ ( -31.10, z-score = -2.16, R) >droSec1.super_0 317179 123 + 21120651 GCGGAUGAGGGCGACUGCUGCUGAUGCUGCUGC-----UGGCCUCUCAAACGAACAAAAAAAAAAAAGGAAGGGUCCAAAACAUCAUUUUGUGUAAUACAAAAAGUAUGUAUGGAAUACAGGGAAUGC------ .((..(((((((.((.((.((....)).)).).-----).))).))))..))...............(((....))).......((((((.(((((((((.......)))))....)))).)))))).------ ( -29.40, z-score = -1.19, R) >droYak2.chr3L 8615963 118 + 24197627 GCAGAUGAGGGCGAAGGCUGCUGAUGCUCC-----------UCUCUCAAA-----AAAGAAAGAGGAGGAAGGGUCCAUAACAUCGUUUUGUGUAAUACAAAAAGUAUGUAUGGAAUACAGGGAAUGCAAAGGA (((......(((....))).(((.(.((((-----------(((.((...-----...)).))))))).)....((((((.((((.(((((((...))))))).).)))))))))...)))....)))...... ( -34.90, z-score = -3.05, R) >droEre2.scaffold_4784 22427255 118 - 25762168 GCAGAUGAGGGCGACGGCUGCUGAUGCUG-----------GCCUCUCAAA-AUCCGAAAAAAAA---GGAAGGGUCCAAAACAUCGUUUUGUGUAAUACAAAAAGUAUGUAUAGAAUACGGGGAAUGCAAAAA- (((..(((((((..((((.......))))-----------))).))))..-.(((.........---(((....)))......((((.((.((((((((.....)))).)))).)).))))))).))).....- ( -30.30, z-score = -1.78, R) >consensus GCAGAUGAGGGCGACGGCUGCUGAUGCUGCUG________GCCUCUCAAACGAACAAAAAAAAA___GGAAGGGUCCAAAACAUCAUUUUGUGUAAUACAAAAAGUAUGUAUGGAAUACAGGGAAUGCAAA___ (((((((.((((..((((.......))))............(((..(....................)..)))))))....)))).((((((((.(((((.......)))))...))))))))..)))...... (-21.35 = -21.19 + -0.16)

| Location | 8,029,873 – 8,030,003 |
|---|---|
| Length | 130 |
| Sequences | 5 |
| Columns | 134 |
| Reading direction | reverse |
| Mean pairwise identity | 83.91 |
| Shannon entropy | 0.27400 |
| G+C content | 0.43319 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
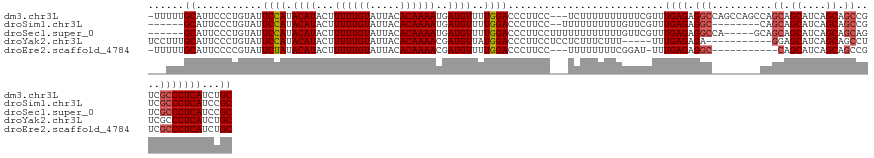
>dm3.chr3L 8029873 130 - 24543557 -UUUUUGCAUUCCCUGUAUUCCAUACAUACUUUUUGUAUUACACAAAAUGAUGUUUUGGACCCUUCC---UCUUUUUUUUUUCGUUUGAGAGGCCAGCCAGCCAGCAGCAUCAGCAGCCGUCGCCCUCAUCUGC -...((((......((((....(((((.......))))))))).....(((((((((((...((.((---((((.............))))))..))....)))).)))))))))))................. ( -24.22, z-score = -0.91, R) >droSim1.chr3L 7493826 118 - 22553184 ------GCAUUCCCUGUAUUCCAUACAUACUUUUUGUAUUACACAAAAUGAUGUUUUGGACCCUUCC--UUUUUUUUUUGUUCGUUUGAGAGGC--------CAGCAGCAUCAGCAGCCGUCGCCCUCAUCCGC ------.............((((.((((...((((((.....))))))..))))..)))).......--.............((..((((.(((--------..((.((....)).))....)))))))..)). ( -25.00, z-score = -2.29, R) >droSec1.super_0 317179 123 - 21120651 ------GCAUUCCCUGUAUUCCAUACAUACUUUUUGUAUUACACAAAAUGAUGUUUUGGACCCUUCCUUUUUUUUUUUUGUUCGUUUGAGAGGCCA-----GCAGCAGCAUCAGCAGCAGUCGCCCUCAUCCGC ------.............((((.((((...((((((.....))))))..))))..))))......................((..((((.(((..-----((.((.((....)).)).)).)))))))..)). ( -26.40, z-score = -2.23, R) >droYak2.chr3L 8615963 118 - 24197627 UCCUUUGCAUUCCCUGUAUUCCAUACAUACUUUUUGUAUUACACAAAACGAUGUUAUGGACCCUUCCUCCUCUUUCUUU-----UUUGAGAGA-----------GGAGCAUCAGCAGCCUUCGCCCUCAUCUGC ......((.....((((..(((((((((.(.((((((.....)))))).)))).))))))......((((((((((...-----...))))))-----------)))).....)))).....)).......... ( -31.50, z-score = -4.81, R) >droEre2.scaffold_4784 22427255 118 + 25762168 -UUUUUGCAUUCCCCGUAUUCUAUACAUACUUUUUGUAUUACACAAAACGAUGUUUUGGACCCUUCC---UUUUUUUUCGGAU-UUUGAGAGGC-----------CAGCAUCAGCAGCCGUCGCCCUCAUCUGC -.....(((....(((........((((.(.((((((.....)))))).)))))...(((....)))---........)))..-..((((.(((-----------..((.......))....)))))))..))) ( -19.30, z-score = -0.18, R) >consensus ___UUUGCAUUCCCUGUAUUCCAUACAUACUUUUUGUAUUACACAAAAUGAUGUUUUGGACCCUUCC___UUUUUUUUUGUUCGUUUGAGAGGC________CAGCAGCAUCAGCAGCCGUCGCCCUCAUCUGC ......((...........((((.((((...((((((.....))))))..))))..)))).............................((((..............((....)).((....))))))....)) (-15.70 = -15.74 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:43 2011