
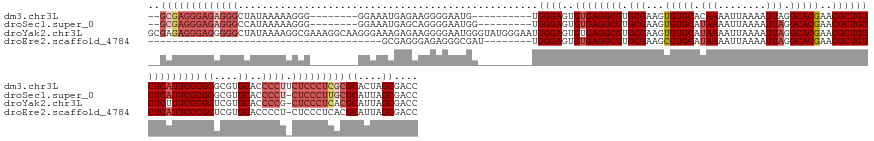
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,017,395 – 8,017,540 |
| Length | 145 |
| Max. P | 0.999714 |

| Location | 8,017,395 – 8,017,540 |
|---|---|
| Length | 145 |
| Sequences | 4 |
| Columns | 165 |
| Reading direction | forward |
| Mean pairwise identity | 76.07 |
| Shannon entropy | 0.37346 |
| G+C content | 0.55967 |
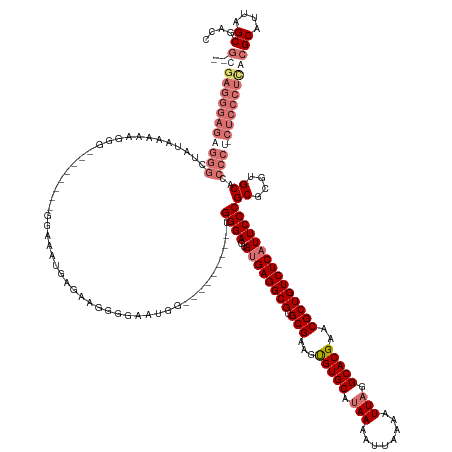
| Mean single sequence MFE | -62.75 |
| Consensus MFE | -40.04 |
| Energy contribution | -40.98 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.39 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8017395 145 + 24543557 --GCGAGGGAGAGGGCUAUAAAAAGGG--------GGAAAUGAGAAGGGGAAUG----------UGGGAGUGUGAGGCGUGCGAAGUGUGCACAAAAUUAAAAUUAGGCACGAACGCUGUCUCAUUCCCGCGCGUGCACCCCUUCUCCCUCGCGCACUAGCGACC --((((((((((...........((((--------(...(((.(...(((((((----------.((.(((((..((((((((.....)))))..(((....)))..)).)..))))).)).))))))).).)))...)))))))))))))))((....)).... ( -60.10, z-score = -3.34, R) >droSec1.super_0 304833 145 + 21120651 --GCGAGGGAGAGGGCCAUAAAAAGGG--------GGAAAUGAGCAGGGGAAUGG---------UGGGAGUGUGAGGCGUGCGAAGUGUGCAUAAAAUUAAAAUUAGGCACGAACGCUGUCUCAUUCCCGCGCGUGCACCCCU-CUCCCUUGCGCAUUAGCGACC --(((((((((((((((((........--------....))).)).((...((((---------(((((..((((((((.(((...(((((.(((........))).)))))..))))))))))))))))).)))...)))))-)))))))))((....)).... ( -64.40, z-score = -4.29, R) >droYak2.chr3L 8603017 164 + 24197627 GCGAGAGGGAGGGGGCUAUAAAAGGCGAAAGGCAAGGGAAAGAGAAGGGGAAUGGGUAUGGGAAUGGGAGUGUGAGGCGUGCGAAGUGUGCAUAAAAUUAAAAUUAGGCACGAACGCUGUCUCUUUCCCGCUCGUGCACCCCG-CUCCCUCACGCAUUAGCGACC (((.(((((((((((........(.(....).)......................(((((((...(((((...((((((.(((...(((((.(((........))).)))))..))))))))).))))).))))))).)))).-))))))).))).......... ( -66.60, z-score = -3.72, R) >droEre2.scaffold_4784 22414454 117 - 25762168 ---------------------------------------GCGAGGGAGAGGGCGAU--------UGGGAGUGUGAGGCGUGCGAAGCGUGCAUAAAAUUAAAAUUAGGCACGAACGCUGUCUCAUUCCCGCUCGUGCACCCCU-CUCCCUCACGCAUUAGCGACC ---------------------------------------..((((((((((((((.--------(((((..((((((((.(((...(((((.(((........))).)))))..)))))))))))))))).))).....))))-))))))).(((....)))... ( -59.90, z-score = -6.21, R) >consensus __GCGAGGGAGAGGGCUAUAAAAAGGG________GGAAAUGAGAAGGGGAAUGG_________UGGGAGUGUGAGGCGUGCGAAGUGUGCAUAAAAUUAAAAUUAGGCACGAACGCUGUCUCAUUCCCGCGCGUGCACCCCU_CUCCCUCACGCAUUAGCGACC .........................................(((.(((((...............((((..((((((((.(((...(((((.(((........))).)))))..)))))))))))))))((....)).))))).))).....(((....)))... (-40.04 = -40.98 + 0.94)
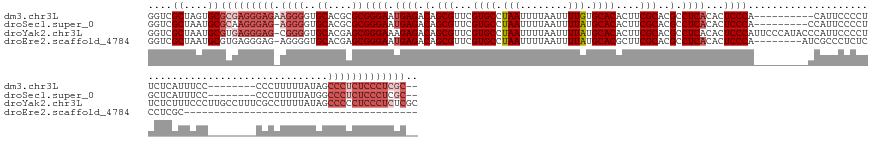
| Location | 8,017,395 – 8,017,540 |
|---|---|
| Length | 145 |
| Sequences | 4 |
| Columns | 165 |
| Reading direction | reverse |
| Mean pairwise identity | 76.07 |
| Shannon entropy | 0.37346 |
| G+C content | 0.55967 |
| Mean single sequence MFE | -51.80 |
| Consensus MFE | -32.99 |
| Energy contribution | -33.17 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 8017395 145 - 24543557 GGUCGCUAGUGCGCGAGGGAGAAGGGGUGCACGCGCGGGAAUGAGACAGCGUUCGUGCCUAAUUUUAAUUUUGUGCACACUUCGCACGCCUCACACUCCCA----------CAUUCCCCUUCUCAUUUCC--------CCCUUUUUAUAGCCCUCUCCCUCGC-- ....((....))((((((((((.((((((....)))((((((((((..(((...((((.(((........))).))))....)))..(.....).......----------.........))))))))))--------............)))))))))))))-- ( -51.10, z-score = -3.22, R) >droSec1.super_0 304833 145 - 21120651 GGUCGCUAAUGCGCAAGGGAG-AGGGGUGCACGCGCGGGAAUGAGACAGCGUUCGUGCCUAAUUUUAAUUUUAUGCACACUUCGCACGCCUCACACUCCCA---------CCAUUCCCCUGCUCAUUUCC--------CCCUUUUUAUGGCCCUCUCCCUCGC-- ....((....))((.((((((-(((((((....)))(((((((((.(((((...((((.(((........))).))))....)))..(.....).......---------.........)))))))))))--------............)))))))))).))-- ( -48.80, z-score = -2.67, R) >droYak2.chr3L 8603017 164 - 24197627 GGUCGCUAAUGCGUGAGGGAG-CGGGGUGCACGAGCGGGAAAGAGACAGCGUUCGUGCCUAAUUUUAAUUUUAUGCACACUUCGCACGCCUCACACUCCCAUUCCCAUACCCAUUCCCCUUCUCUUUCCCUUGCCUUUCGCCUUUUAUAGCCCCCUCCCUCUCGC ..........(((.(((((((-.(((((((....))((((((((((..(((...((((.(((........))).))))....)))..(.....)..........................))))))))))...................)))))))))))).))) ( -54.90, z-score = -4.13, R) >droEre2.scaffold_4784 22414454 117 + 25762168 GGUCGCUAAUGCGUGAGGGAG-AGGGGUGCACGAGCGGGAAUGAGACAGCGUUCGUGCCUAAUUUUAAUUUUAUGCACGCUUCGCACGCCUCACACUCCCA--------AUCGCCCUCUCCCUCGC--------------------------------------- ....((....))(((((((((-((((..((....))((((.((((.(.(((..(((((.(((........))).)))))...)))..).))))...)))).--------....)))))))))))))--------------------------------------- ( -52.40, z-score = -4.45, R) >consensus GGUCGCUAAUGCGCGAGGGAG_AGGGGUGCACGAGCGGGAAUGAGACAGCGUUCGUGCCUAAUUUUAAUUUUAUGCACACUUCGCACGCCUCACACUCCCA_________CCAUUCCCCUUCUCAUUUCC________CCCUUUUUAUAGCCCUCUCCCUCGC__ ...(((....))).....(((.(((((.((....))((((.((((.(.(((...((((.(((........))).))))....)))..).))))...))))...............))))).)))......................................... (-32.99 = -33.17 + 0.19)
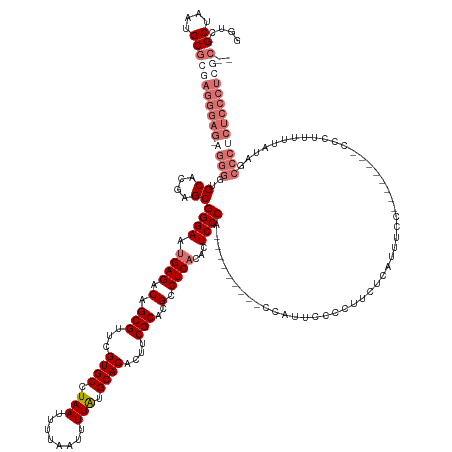
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:40 2011