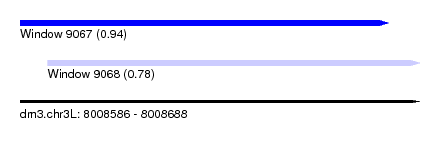
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 8,008,586 – 8,008,688 |
| Length | 102 |
| Max. P | 0.941887 |

| Location | 8,008,586 – 8,008,680 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.28 |
| Shannon entropy | 0.26867 |
| G+C content | 0.29010 |
| Mean single sequence MFE | -14.08 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.941887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
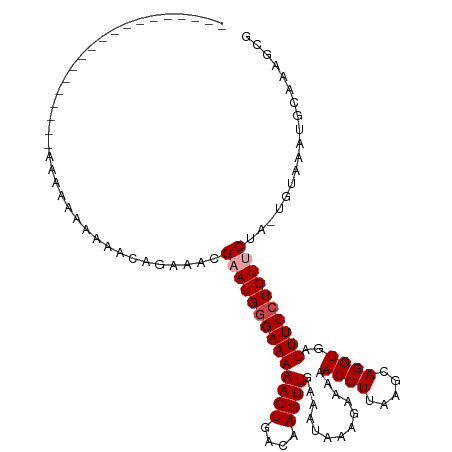
>dm3.chr3L 8008586 94 + 24543557 ------------------AAAUGUAGAAAAAAAAAAAC----GAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCUA-UGUAAAU ------------------....................----.....((((((((((((((....)))).............((((.....))))..))))))))))..-....... ( -15.60, z-score = -2.48, R) >droSim1.chr3L 7474483 98 + 22553184 ------------------AAAUGUAGAAAAAAAAAAAAAACAGAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUCCUA-UGUAAAU ------------------...............................((((((..((((....))))........((((.((((.....))))..))))...)))))-)...... ( -12.60, z-score = -1.22, R) >droSec1.super_0 300575 92 + 21120651 ------------------AAAUGUAAAAAAAAAACA------GAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUCCUA-UGUAAAU ------------------..................------.......((((((..((((....))))........((((.((((.....))))..))))...)))))-)...... ( -12.60, z-score = -1.23, R) >droYak2.chr3L 8596985 97 + 24197627 ------------------AAAUGUAAAAGAAAAAAAAUAA-AGAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCUA-UGUAAAU ------------------......................-......((((((((((((((....)))).............((((.....))))..))))))))))..-....... ( -15.60, z-score = -2.71, R) >droEre2.scaffold_4784 22410253 93 - 25762168 ------------------AAAUGUAGAAAAAAAAACA-----GAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCUA-UGUAAAU ------------------...................-----.....((((((((((((((....)))).............((((.....))))..))))))))))..-....... ( -15.60, z-score = -2.25, R) >droAna3.scaffold_13337 7277895 114 - 23293914 GAAACGUACAAAACUGAAAAAGCUAGAAAACUAAAAAGC---AGAAAGAAUGAGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCCAACUUCUAU .............(((.......(((....))).....)---))....................((((((.....(((......))).((((((......)))))).)))))).... ( -12.50, z-score = 0.30, R) >consensus __________________AAAUGUAGAAAAAAAAAAA_____GAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCUA_UGUAAAU ...............................................((((((((((((((....)))).............((((.....))))..)))))))))).......... (-13.02 = -13.52 + 0.50)
| Location | 8,008,593 – 8,008,688 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.17 |
| Shannon entropy | 0.25922 |
| G+C content | 0.32230 |
| Mean single sequence MFE | -14.64 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
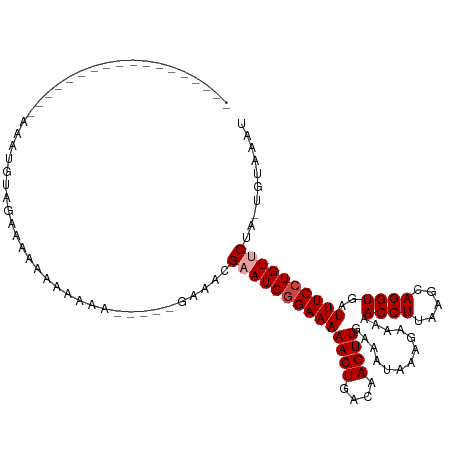
>dm3.chr3L 8008593 95 + 24543557 ------------------GAAAAAAAAAAACGAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCUA-UGUAAAUGCAAAGCG ------------------..................((((((((((((((....)))).............((((.....))))..))))))))))..-(((....)))..... ( -15.70, z-score = -1.98, R) >droSim1.chr3L 7474490 99 + 22553184 --------------GAAAAAAAAAAAAAACAGAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUCCUA-UGUAAAUGCAAAGCG --------------........................((((((..((((....))))........((((.((((.....))))..))))...)))))-).............. ( -12.60, z-score = -0.74, R) >droSec1.super_0 300581 94 + 21120651 -------------------AAAAAAAAAACAGAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUCCUA-UGUAAAUGCAAAGCG -------------------...................((((((..((((....))))........((((.((((.....))))..))))...)))))-).............. ( -12.60, z-score = -0.65, R) >droYak2.chr3L 8596992 98 + 24197627 ---------------AAAGAAAAAAAAUAAAGAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCUA-UGUAAAUGCAAAGCG ---------------.....................((((((((((((((....)))).............((((.....))))..))))))))))..-(((....)))..... ( -15.70, z-score = -1.91, R) >droEre2.scaffold_4784 22410260 94 - 25762168 -------------------GAAAAAAAAACAGAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCUA-UGUAAAUGCAAAGCG -------------------.................((((((((((((((....)))).............((((.....))))..))))))))))..-(((....)))..... ( -15.70, z-score = -1.84, R) >droAna3.scaffold_13337 7277903 113 - 23293914 CAAAACUGAAAAAGCUAGAAAACUAAAAAGCAGAAAGAAUGAGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCCAACUUCUAUGCAAAGU- .............(((((((.........((((...............))).)..((((.....(((......))).((((((......)))))).))))))))).)).....- ( -15.56, z-score = -0.38, R) >consensus ___________________AAAAAAAAAACAGAAACGAAUGGGAAAAACUGACAAGUUGAAAUAAAGAAAAACCUUAAGCAGGUGAUUUCCUGUUCUA_UGUAAAUGCAAAGCG ....................................((((((((((((((....)))).............((((.....))))..)))))))))).................. (-13.02 = -13.52 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:38 2011