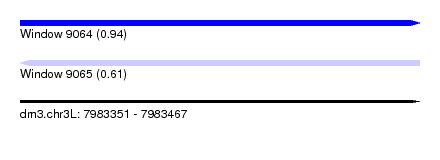
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,983,351 – 7,983,467 |
| Length | 116 |
| Max. P | 0.936585 |

| Location | 7,983,351 – 7,983,467 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.55 |
| Shannon entropy | 0.05979 |
| G+C content | 0.47142 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -34.14 |
| Energy contribution | -34.78 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
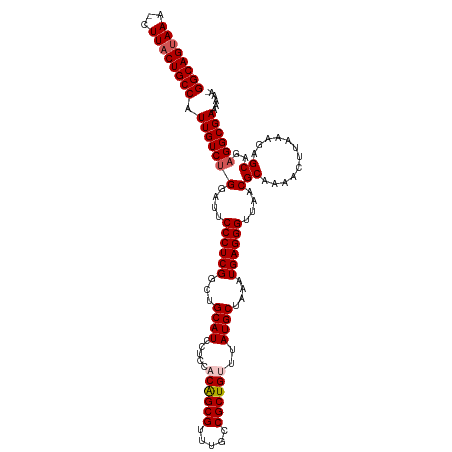
>dm3.chr3L 7983351 116 + 24543557 GGCAGUAAA-CUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUAAAGAGCAGAGGCGAAAAAAA (((((((..-..))))))).(((((((....((((((...((((.....((((((.....))))))..))))....))))))....)((............))..))))))...... ( -37.60, z-score = -2.33, R) >droSim1.chr3L 7455595 115 + 22553184 GGCAGUAAA-CUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUAAAGAGCAGAGGCGAAAAAA- (((((((..-..))))))).(((((((....((((((...((((.....((((((.....))))))..))))....))))))....)((............))..)))))).....- ( -37.60, z-score = -2.35, R) >droSec1.super_0 275774 116 + 21120651 GGCAGAAAAACUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUAAAGAGCAGAGGCGAAAAAA- (((((.........))))).(((((((....((((((...((((.....((((((.....))))))..))))....))))))....)((............))..)))))).....- ( -34.10, z-score = -1.35, R) >droYak2.chr3L 8570949 115 + 24197627 GGCAGUAAA-CUUACUGCCAUUGUCUGGAUUCCCUCGACUGCAUCCUCCUCGGCGUCUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUAAAGAGCAGAGGCGAAAAAA- (((((((..-..))))))).(((((((....((((((...((((...(..((((....))))..)...))))....))))))....)((............))..)))))).....- ( -35.40, z-score = -1.96, R) >droEre2.scaffold_4784 22385345 115 - 25762168 GGCAGUAAA-CUUACUGCCAUUGUCCGGAUUCCCUCGACUGCAUCCUCCUCAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUGAAGAGCAGAGGCGAAAAAA- (((((((..-..))))))).(((((((....((((((...((((......(((((.....)))))...))))....))))))....)((............)).).))))).....- ( -34.50, z-score = -1.68, R) >consensus GGCAGUAAA_CUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUAAAGAGCAGAGGCGAAAAAA_ (((((((.....))))))).(((((((....((((((...((((.....((((((.....))))))..))))....))))))....)((............))..))))))...... (-34.14 = -34.78 + 0.64)

| Location | 7,983,351 – 7,983,467 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 96.55 |
| Shannon entropy | 0.05979 |
| G+C content | 0.47142 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -32.08 |
| Energy contribution | -32.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.610985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
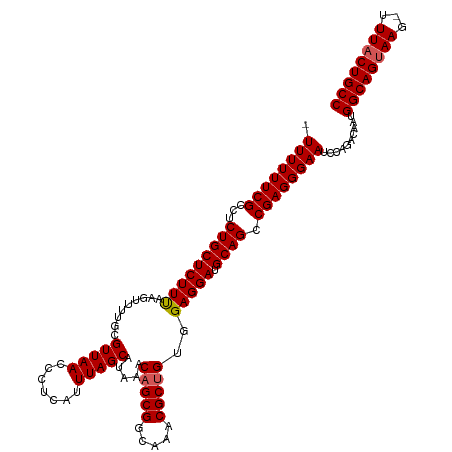
>dm3.chr3L 7983351 116 - 24543557 UUUUUUUCGCCUCUGCUCUUUAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAG-UUUACUGCC ...((..(((....(((.....)))...)))..))(((((.....((((..((((((.....)))))).....))))....)))))............(((((((..-..))))))) ( -34.70, z-score = -1.38, R) >droSim1.chr3L 7455595 115 - 22553184 -UUUUUUCGCCUCUGCUCUUUAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAG-UUUACUGCC -..((..(((....(((.....)))...)))..))(((((.....((((..((((((.....)))))).....))))....)))))............(((((((..-..))))))) ( -34.70, z-score = -1.44, R) >droSec1.super_0 275774 116 - 21120651 -UUUUUUCGCCUCUGCUCUUUAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAGUUUUUCUGCC -..((..(((....(((.....)))...)))..))(((((.....((((..((((((.....)))))).....))))....)))))............(((((.........))))) ( -31.60, z-score = -0.41, R) >droYak2.chr3L 8570949 115 - 24197627 -UUUUUUCGCCUCUGCUCUUUAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAGACGCCGAGGAGGAUGCAGUCGAGGGAAUCCAGACAAUGGCAGUAAG-UUUACUGCC -....(((.((((.(((.....))).(((((((...((((.....((.......))(((....)))))))..)))))))..)))))))..........(((((((..-..))))))) ( -35.20, z-score = -1.71, R) >droEre2.scaffold_4784 22385345 115 + 25762168 -UUUUUUCGCCUCUGCUCUUCAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGAGGAGGAUGCAGUCGAGGGAAUCCGGACAAUGGCAGUAAG-UUUACUGCC -((((((((...(((((((((.........(((((.......))))).....(((((.....)))))..))))).)))).))))))))..........(((((((..-..))))))) ( -36.30, z-score = -1.80, R) >consensus _UUUUUUCGCCUCUGCUCUUUAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAG_UUUACUGCC .((((((((...(((((((((.........(((((.......))))).....(((((.....)))))..))))).)))).))))))))..........(((((((.....))))))) (-32.08 = -32.32 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:35 2011