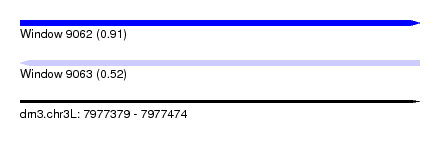
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,977,379 – 7,977,474 |
| Length | 95 |
| Max. P | 0.912295 |

| Location | 7,977,379 – 7,977,474 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.30 |
| Shannon entropy | 0.13207 |
| G+C content | 0.35042 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -21.80 |
| Energy contribution | -21.36 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
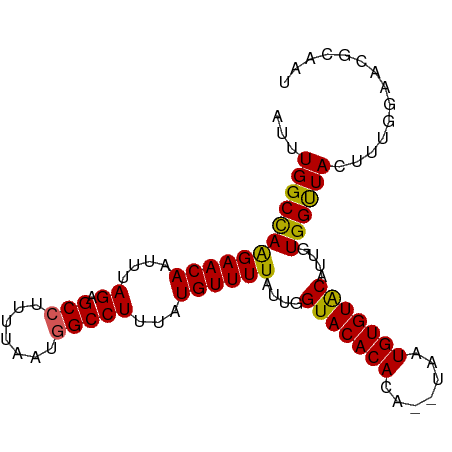
>dm3.chr3L 7977379 95 + 24543557 AUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUAGCCUUUAUGUUUUAUUGGUACACACACAUAAUGUGUACAUUGUGGUUACUUUGGAACGCAAU ...((((((((((((...(((((.((......)).)))))))))))....(((((((.......)))))))....)))))).............. ( -19.30, z-score = -0.66, R) >droEre2.scaffold_4784 22381795 89 - 25762168 AUUUGGCCAGGAACAGUUUAGAGCCUUU-AGUGGCCUUUAUGUUUUAUUGGUACACAUG----AUGUGUGCAUUGUGGCUACAUUG-AACGCAAU ...((((((((((((....((.(((...-...)))))...))))))....((((((...----..))))))....)))))).....-........ ( -26.00, z-score = -1.64, R) >droYak2.chr3L 8567263 90 + 24197627 AUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUGGUACACAUA----AUGUGUACAUUGUGGUUACUUUG-AACGCAAU ...((((((((((((....((.(((.......)))))...))))))....((((((...----..))))))....)))))).....-........ ( -22.00, z-score = -1.89, R) >droSec1.super_0 272126 93 + 21120651 AUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUGGUACACACA--UAAUGUGUACAUUGUGGUUACUUUGGAACGCAAU ...((((((((((((....((.(((.......)))))...))))))....(((((((..--...)))))))....)))))).............. ( -22.40, z-score = -1.57, R) >droSim1.chr3L 7452021 93 + 22553184 AUUUGGCUAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUGGUACACACA--UAAUGUGUACAUUGUGGUUACUUUGGAACGCAAU .....((........((..((.(((.......)))))..))((((((..((((.((.((--(((((....))))))))))))).))))))))... ( -21.10, z-score = -1.40, R) >consensus AUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUGGUACACACA__UAAUGUGUACAUUGUGGUUACUUUGGAACGCAAU ...((((((((((((....((.(((.......)))))...))))))....(((((((.......)))))))....)))))).............. (-21.80 = -21.36 + -0.44)

| Location | 7,977,379 – 7,977,474 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.30 |
| Shannon entropy | 0.13207 |
| G+C content | 0.35042 |
| Mean single sequence MFE | -16.66 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.10 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.521721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
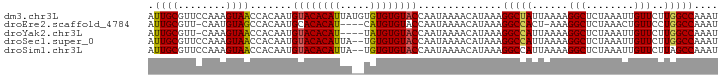
>dm3.chr3L 7977379 95 - 24543557 AUUGCGUUCCAAAGUAACCACAAUGUACACAUUAUGUGUGUGUACCAAUAAAACAUAAAGGCUAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAU .((((........)))).......((((((((.....))))))))..............(((((....(..((........))..)))))).... ( -17.00, z-score = -0.37, R) >droEre2.scaffold_4784 22381795 89 + 25762168 AUUGCGUU-CAAUGUAGCCACAAUGCACACAU----CAUGUGUACCAAUAAAACAUAAAGGCCACU-AAAGGCUCUAAACUGUUCCUGGCCAAAU ..(((((.-..)))))((((...((((((...----..)))))).......((((...(((((...-...))).))....))))..))))..... ( -16.00, z-score = -0.10, R) >droYak2.chr3L 8567263 90 - 24197627 AUUGCGUU-CAAAGUAACCACAAUGUACACAU----UAUGUGUACCAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAU .((((...-....)))).......((((((..----...))))))..............(((((....(..((........))..)))))).... ( -16.50, z-score = -1.05, R) >droSec1.super_0 272126 93 - 21120651 AUUGCGUUCCAAAGUAACCACAAUGUACACAUUA--UGUGUGUACCAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAU .((((........)))).......((((((((..--.))))))))..............(((((....(..((........))..)))))).... ( -17.90, z-score = -1.05, R) >droSim1.chr3L 7452021 93 - 22553184 AUUGCGUUCCAAAGUAACCACAAUGUACACAUUA--UGUGUGUACCAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUAGCCAAAU .((((........))))..(((((((((((((..--.)))))))).............(((((.......))).))..)))))............ ( -15.90, z-score = -1.02, R) >consensus AUUGCGUUCCAAAGUAACCACAAUGUACACAUUA__UGUGUGUACCAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAU .((((........)))).......((((((((.....))))))))..............(((((.....((........)).....))))).... (-16.38 = -16.10 + -0.28)
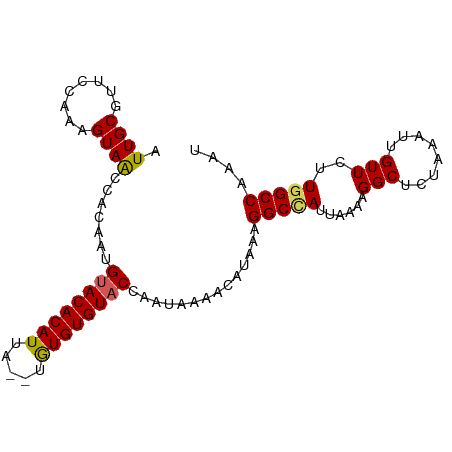
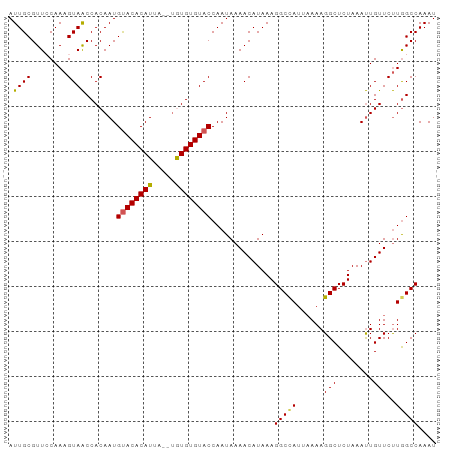
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:34 2011