| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,948,692 – 7,948,753 |
| Length | 61 |
| Max. P | 0.615354 |

| Location | 7,948,692 – 7,948,753 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 71.61 |
| Shannon entropy | 0.48758 |
| G+C content | 0.44301 |
| Mean single sequence MFE | -16.62 |
| Consensus MFE | -7.00 |
| Energy contribution | -7.76 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
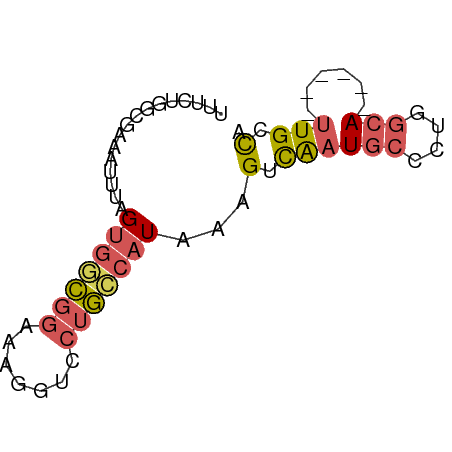
>dm3.chr3L 7948692 61 - 24543557 UUUCUGGCGAAAUUUAGUGGCGGAAAACUCCUGCCAUAAAGUCAAUGCCCUUGCA------UUGCCA .....(((...((((.((((((((....))).))))).)))).(((((....)))------))))). ( -19.20, z-score = -2.47, R) >droSim1.chr3L 7423411 61 - 22553184 UUUCUGGCGAAAUUUAGUGGCGGAAAGGUCCUGCCAUAAAGUCAAUGCCCUGGCA------UUGCCA .....(((...((((.(((((((.......))))))).)))).(((((....)))------))))). ( -21.20, z-score = -1.89, R) >droSec1.super_0 242731 61 - 21120651 UUUCUGACGAAAUUUAGUGGCGGAAAGGUCCUGCCAUAAAGUCAAUGCCCUGGCA------UUGCCA ((((....))))....(((((((.......)))))))...(.((((((....)))------))).). ( -18.30, z-score = -1.59, R) >droEre2.scaffold_4784 22351530 61 + 25762168 UUUCCGUCGAAAUUUAGUGUCGGAAAGGUCCUGCCAUAAAGUCAAUGCCCUGGCA------UUGCCA ((((((.((........)).))))))((.....)).....(.((((((....)))------))).). ( -17.20, z-score = -2.32, R) >droWil1.scaffold_181089 11925032 67 - 12369635 CUUAUCCCGUUAUUGCGUGAUACUUAAGUCUAAUAUUAUCGUUGUUUUUUUUUCAAGCAAUUUGCUG ........((.(((((.(((.....(((..((((......))))..)))...))).)))))..)).. ( -7.20, z-score = -0.82, R) >consensus UUUCUGGCGAAAUUUAGUGGCGGAAAGGUCCUGCCAUAAAGUCAAUGCCCUGGCA______UUGCCA ...........((((.(((((((.......))))))).))))........(((((.......))))) ( -7.00 = -7.76 + 0.76)

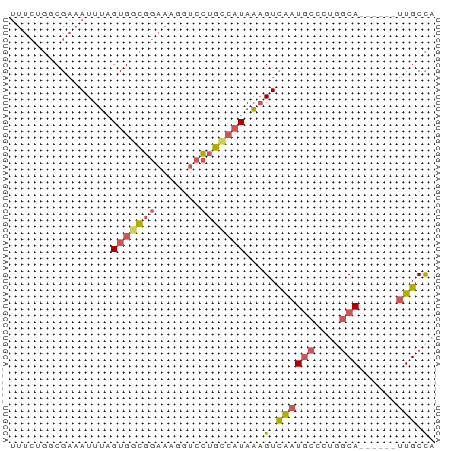
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:28 2011