| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,932,887 – 7,932,981 |
| Length | 94 |
| Max. P | 0.922323 |

| Location | 7,932,887 – 7,932,981 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.91 |
| Shannon entropy | 0.52980 |
| G+C content | 0.50544 |
| Mean single sequence MFE | -24.19 |
| Consensus MFE | -13.71 |
| Energy contribution | -14.04 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
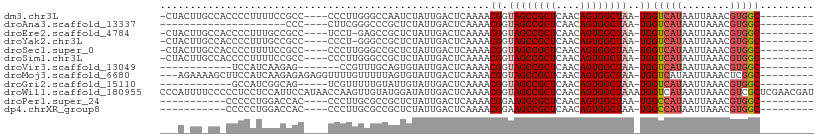
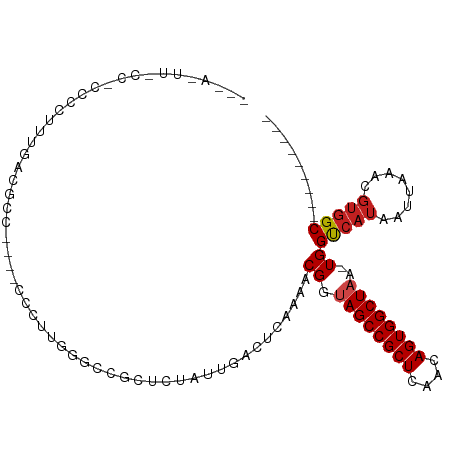
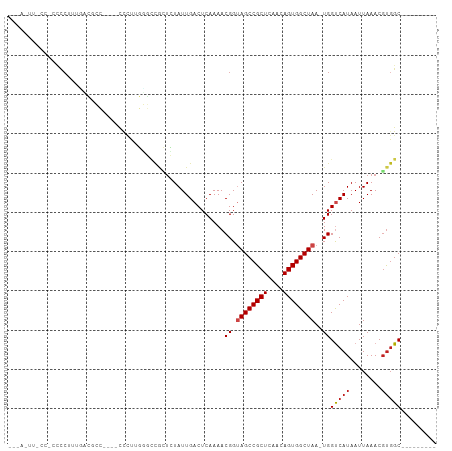
>dm3.chr3L 7932887 94 + 24543557 -CUACUUGCCACCCCUUUUCCGCC----CCCUUGGGCCAAUCUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACGUGGC--------- -......(((((.........(((----(....))))........((((.((......((((((((....)))))))).-)))))).........)))))--------- ( -28.00, z-score = -2.48, R) >droAna3.scaffold_13337 7205981 74 - 23293914 ---------------------CCC----CUUCGGGCCCGCUCUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACGUGGC--------- ---------------------..(----(....)).((((.....((((.((......((((((((....)))))))).-)))))).........)))).--------- ( -20.14, z-score = -0.95, R) >droEre2.scaffold_4784 22335117 93 - 25762168 -CUACUUGCCACCCCUUUGCCGCC----UCCU-GAGCCGCUCUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACGUGGC--------- -......(((((((.((.(((((.----...(-((((.((((..((.......))..).))).)))))...))))).))-.))............)))))--------- ( -26.10, z-score = -1.93, R) >droYak2.chr3L 8521010 93 + 24197627 -CUACUUGCCACCCCUUUGCCGCC----CCCU-GGGCCGCUCUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACGUGGC--------- -......(((((......((.(((----(...-)))).)).....((((.((......((((((((....)))))))).-)))))).........)))))--------- ( -30.20, z-score = -2.30, R) >droSec1.super_0 226951 94 + 21120651 -CUACUUGCCACCCCUUUUCCGCC----CCCUUGGGCCGCUCUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACGUGGC--------- -......(((((.........(((----(....))))........((((.((......((((((((....)))))))).-)))))).........)))))--------- ( -28.00, z-score = -2.07, R) >droSim1.chr3L 7406803 94 + 22553184 -CUACUUGCCACCCCUUUUCCGCC----CCCUUGGGCCGCUCUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACGUGGC--------- -......(((((.........(((----(....))))........((((.((......((((((((....)))))))).-)))))).........)))))--------- ( -28.00, z-score = -2.07, R) >droVir3.scaffold_13049 4482737 80 - 25233164 ------------UCCAUCAAGAG-------CCGUUUGCAGUGUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACGUGGC--------- ------------.((((.....(-------((((((..(((......)))..)))))))(((((((....)))))))..-...............)))).--------- ( -21.90, z-score = -1.55, R) >droMoj3.scaffold_6680 16304334 96 - 24764193 ---AGAAAAGCUUCCAUCAAGAGAGAGGUUUUGUUUUUAGUGUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACUCGGC--------- ---...(((((((((.......).))))))))(..((((((....((((.((......((((((((....)))))))).-))))))..))))))..)...--------- ( -22.90, z-score = -1.04, R) >droGri2.scaffold_15110 11781223 82 + 24565398 ------------GCCAUCGGCAG-----UCGUUUUUGUAUUGUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA-UGGUCAUAAUUAAACGUGGC--------- ------------((((.((((((-----(((....)).)))))..((((.((......((((((((....)))))))).-))))))........))))))--------- ( -23.70, z-score = -1.99, R) >droWil1.scaffold_180955 162944 109 - 2875958 CCCAUUUUCCCCCUCCUCCAUUCCAUAACCAAGUUGUAUGGAUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAAAUGGUCAUAAUUAAACGUCGCUCGAACGAU .(((((............((.((((((.........))))))...))...........((((((((....)))))))))))))............((((......)))) ( -21.30, z-score = -0.93, R) >droPer1.super_24 61089 84 - 1556852 -----------CCCCCUGGACCAC----CCCUUGCGCCGCUCUAUUGACUCAAAACGGAAGCCGCUCAACAGUGGCUAA-UGGCCAUAAUUAAACGUGGC--------- -----------.....(((.((((----...(((.((.((((..((.......))..).))).)).)))..))))))).-..(((((........)))))--------- ( -20.00, z-score = -0.18, R) >dp4.chrXR_group8 1557848 84 + 9212921 -----------CCCCCUGGACCAC----CCCUUGCGCCGCUCUAUUGACUCAAAACGGAAGCCGCUCAACAGUGGCUAA-UGGCCAUAAUUAAACGUGGC--------- -----------.....(((.((((----...(((.((.((((..((.......))..).))).)).)))..))))))).-..(((((........)))))--------- ( -20.00, z-score = -0.18, R) >consensus ___A_UU_CC_CCCCUUUGACGCC____CCCUUGGGCCGCUCUAUUGACUCAAAACGGUAGCCGCUCAACAGUGGCUAA_UGGUCAUAAUUAAACGUGGC_________ .............................................((((.((......((((((((....))))))))..))))))....................... (-13.71 = -14.04 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:27 2011