| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,930,068 – 7,930,173 |
| Length | 105 |
| Max. P | 0.728365 |

| Location | 7,930,068 – 7,930,173 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 63.29 |
| Shannon entropy | 0.67558 |
| G+C content | 0.42957 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -8.81 |
| Energy contribution | -8.32 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
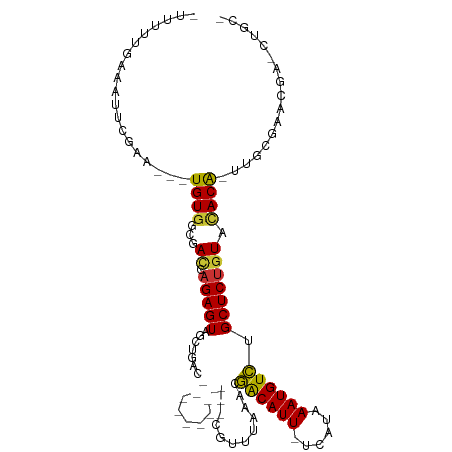
>dm3.chr3L 7930068 105 + 24543557 GUUUUUGACAUUCGAA---UGUGGCGAUCAGAGUAGCUGCUGCAUUUAGCCUGCUUUUAAACGACAUU-UUAUAAAUGUCUGCUCUGUACACG-UUGCGAAUGAACUGCG ........((((((((---((((.....(((((((((.((((....))))..))........((((((-.....)))))))))))))..))))-)).))))))....... ( -32.70, z-score = -2.94, R) >droSec1.super_0 224147 104 + 21120651 -GUUCUGACAUUCGGA---UGUCGCGAUCAGAGUAGCUGCUGCAUUUAGCCUGCUUUUAAACGACAUU-UCAUAAAUGUCUGCUCUGUACACG-UUGCGAAUGAACUGCG -((((.(((((....)---))))((((((((((((((.((((....))))..))........((((((-.....))))))))))))).....)-))))....)))).... ( -30.20, z-score = -1.62, R) >droYak2.chr3L 8518194 105 + 24197627 GUUUUUGAAAUUCGAA---UGUGGCGAUCAGAGUAGCUGCUGCAUUUAGCAUGCUUUUAAACGACAUU-UCAUAAAUGUCUGCUCUGUACACG-UUGCGAAAGAGCUGCG ((((((.....(((((---((((.....((((((((((((((....))))).))........((((((-.....)))))))))))))..))))-)).))))))))).... ( -30.80, z-score = -1.38, R) >droEre2.scaffold_4784 22332408 105 - 25762168 GUUUUUGAAAUUCGAA---UGUGGCGAUCAGAGUAGCUGCUGCAUUUAGCAUGCUUUUAAACGACAUU-UCAUAAAUGUCUGCUCUGUACACG-UUGCGAAUGAACUGCA ((.......(((((((---((((.....((((((((((((((....))))).))........((((((-.....)))))))))))))..))))-)).))))).....)). ( -31.60, z-score = -2.16, R) >droAna3.scaffold_13337 7203451 90 - 23293914 --UGUCCCAGUUCGAA---UGUGGCGACCAGAGUAGUUGAC------------UGUUUAAACGACAUU-UCGCCAAUGUCGGCUCUGUGCACA-UUCCGAAC-ACUUGCU --.......(((((((---((((.((..((((((((....)------------).......(((((((-.....)))))))))))))))))))-)).)))))-....... ( -27.90, z-score = -2.06, R) >droVir3.scaffold_13049 20747545 86 + 25233164 ---CAAAGGCGGCUAAGCAUGUGGCAACAAGAGUAGUU-AC------------UGUUUAAAUAACAUU-UUAUAAAUGUUAGCUCUGUGCACAGCAGCCAGCC------- ---....(((((((..((.((((.((...(((((..((-(.------------....))).(((((((-.....)))))))))))).)))))))))))).)))------- ( -25.00, z-score = -1.61, R) >droMoj3.scaffold_6680 8456913 87 + 24764193 CGCGGCAGGCUACGA----UGUGGUAACAAGAGUAGUUUAC------------UGUUUAAAUAGCAUUGUUAUAAAUGUUAGCUCUGUGCACAGCAGCCAGCC------- ...(((.((((....----((((...(((.((((((....)------------).......(((((((......)))))))))))))).))))..)))).)))------- ( -24.90, z-score = -0.74, R) >droGri2.scaffold_15110 12782092 87 - 24565398 CAUUGCCAGCUGUUAAGCGUGUGUUAACACGAGUAGUU-AC------------UGUUUAAAUAACAUU-UUAUAAAUGUUAGCUCC-UGUACA-CAGCCAGCC------- ....((..(((((((..(((((....)))))..))((.-((------------........(((((((-.....))))))).....-.)))))-))))..)).------- ( -19.44, z-score = -0.92, R) >consensus _UUUUUGAAAUUCGAA___UGUGGCGACCAGAGUAGCUGAC____________CGUUUAAACGACAUU_UCAUAAAUGUCUGCUCUGUACACA_UUGCGAACGA_CUGC_ ...................((((...((.(((((.......((.....))............((((((......)))))).))))))).))))................. ( -8.81 = -8.32 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:26 2011