
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,929,920 – 7,930,015 |
| Length | 95 |
| Max. P | 0.676428 |

| Location | 7,929,920 – 7,930,015 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 63.70 |
| Shannon entropy | 0.72737 |
| G+C content | 0.46606 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -9.35 |
| Energy contribution | -10.50 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.676428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
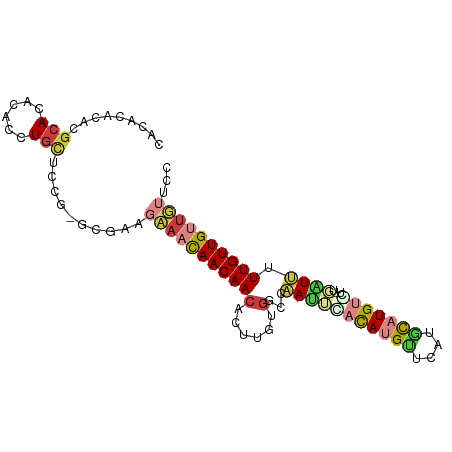
>dm3.chr3L 7929920 95 - 24543557 CACACACAAGCACACACCUGCUCCG-GCGAAGCAACAACAACAUUCGUGGCCGAUUCAUAUGCUCAUGCAUGUGCUAUGAUCUUUGUUGUUGUUCC ..(.(...((((......))))...-).).(((((((((((...(((((((......((((((....)))))))))))))...))))))))))).. ( -28.70, z-score = -2.32, R) >droSim1.chr3L 7403471 96 - 22553184 CACACACGCGCACACACCUGCUCCGCGCGAAGCAACAACAACACUUGGGGCCGAUUCACAUGCUCAUGCAUGUGCUAUGAUCUUUGUUGUUGUUCC ......(((((.............)))))....((((((((((...(((..((...(((((((....)))))))...))..))))))))))))).. ( -31.52, z-score = -2.46, R) >droSec1.super_0 224012 84 - 21120651 ----CACACGCACACA-CUGCUGCGCGCGAAGCAACAACAACACUUGUGGCCGAUUCACAUGCUCAUGCAUG-------AUCUUUGUUGUUGUUCC ----..(.(((.((..-....)).))).).(((((((((((((....)).........(((((....)))))-------....))))))))))).. ( -23.60, z-score = -0.91, R) >droYak2.chr3L 8518047 91 - 24197627 CACACACACGCACACACCUGCUUC----GAUGAAACAACAACACUUGUGGCCAAUUUACAUGUGCAUGCAUGUGCUGUG-UUUUUGUUGUUUUUUC .........(((......)))...----((.((((((((((....((..((......((((((....))))))))..))-...)))))))))).)) ( -19.30, z-score = 0.71, R) >droEre2.scaffold_4784 22332261 91 + 25762168 CACACACACGCACACACCUGCUCC----GACGAAACAACAACGCUUGCGGCUAAUUUACAUGUGCAUGCAUGU-CUGUGAUUUUUGUUGUUUUUUC .........(((......)))...----((.((((((((((.(.(..(((.......((((((....))))))-)))..).).)))))))))).)) ( -20.31, z-score = -0.61, R) >droAna3.scaffold_13337 7203318 90 + 23293914 CACAGGC-UGCAUCCAUCUGUCCCA-ACAAAGAAAUAACAACUUUUGUGUCUCAUUUACAUGUGUGCUU-UGCAGUUGUGUUGUUGUUGGCUG--- .(((((.-((....))))))).(((-((((...((((.(((((.....((..(((......))).))..-...))))))))).)))))))...--- ( -18.70, z-score = 0.63, R) >droGri2.scaffold_15110 12781970 83 + 24565398 CACGGCGAUCAAUCCGCAUACUGGAUAUCGAAUACCAACAACUUCCA----CAACACACA-GUUCACAUUUGU-----UGUUGUUGUUGUUCG--- ...((((((..(((((.....)))))))))....))((((((....(----(((((.(((-(.......))))-----)))))).))))))..--- ( -18.20, z-score = -1.37, R) >consensus CACACACACGCACACACCUGCUCCG_GCGAAGAAACAACAACACUUGUGGCCAAUUCACAUGUUCAUGCAUGU_CU_UGAUUUUUGUUGUUGUUCC .........(((......)))..........((((((((((...(((....)))..(((((((....))))))).........))))))))))... ( -9.35 = -10.50 + 1.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:26 2011