
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,923,103 – 7,923,204 |
| Length | 101 |
| Max. P | 0.979894 |

| Location | 7,923,103 – 7,923,204 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.60041 |
| G+C content | 0.62711 |
| Mean single sequence MFE | -44.48 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.11 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7923103 101 + 24543557 AGGGCUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUC-CGAGACCGUCCGCAACGCG-UUCCGUUGGGCGGUUGGAGGGGCUGUCCAUAUAGAAAGGUU------- .((((.......((((((((......)))))))).((((((-(..(((((.((.(((((..-...)))))))))))))))))))..)))).............------- ( -48.60, z-score = -2.58, R) >droSim1.chr3L 7396629 102 + 22553184 AGGGCUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUC-CGAGACCGUCCGCAACGCG-UUCCGUUGGGCGGUUGGAGGGGCUGUCAGGGGGUGAAAGGUU------ ..((((((((((((((((((......)))))))).((((((-(..(((((.((.(((((..-...)))))))))))))))))))......)))))....)))))------ ( -51.80, z-score = -1.89, R) >droSec1.super_0 217148 102 + 21120651 AGGGCUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUC-CGAGACCGUCCGCAACGCG-UUCCGUUGGGCGGUUGGAGGGGCUGUCAGGGGGUGAAAGGUU------ ..((((((((((((((((((......)))))))).((((((-(..(((((.((.(((((..-...)))))))))))))))))))......)))))....)))))------ ( -51.80, z-score = -1.89, R) >droYak2.chr3L 8511048 102 + 24197627 AGGACUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUC-CGAGACCGUCCGCAACGCG-UUCCGUUGGGCGGUUGGAGGGGCUGUCCAUAUAGAAGCGGUU------ .((((.......((((((((......)))))))).((((((-(..(((((.((.(((((..-...)))))))))))))))))))..))))..............------ ( -49.20, z-score = -2.76, R) >droEre2.scaffold_4784 22325353 101 - 25762168 AGGACUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUC-CGAGACCGUCCGCAACGCG-UUCCGUUGGGCGGCUGGAGGAGCUGUUCAUAUAGA-GCGGUA------ .(((.....)))((((((((......))))))))...((((-(.((.(((.((.(((((..-...))))))))))))))))).(((((((.....))-))))).------ ( -46.80, z-score = -2.60, R) >droAna3.scaffold_13337 7197046 98 - 23293914 AGGGCUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUC-CGAGACCGUCCGCGACGCG-UUCCGUCGGGCGGUUGGAGGGGCUGUUUGGAAGAAGUU---------- ..((((((.(((((((((((......)))))))).((((((-(..(((((((((((.....-...)).))))))))))))))))......))).))))))---------- ( -55.30, z-score = -3.72, R) >dp4.chrXR_group8 7645113 108 - 9212921 AGGACUUCCUCCCCCGGUAUUACGAAACAUCGGCCCCCCUC-CCAGCCCGUCGGCGAUGCG-UUCCGUCGGGCGAUUGGAGGGGCUGUCACGAAAUAGAGAAGAGGCAUU .......((((...(..((((.((......(((((((((..-...(((((.(((.......-..))).)))))....)).)))))))...)).))))..)..)))).... ( -38.50, z-score = 0.10, R) >droPer1.super_24 1485873 108 + 1556852 AGGACUUCCUCCCCCGGUAUUACGAAACAUCGGCCCCCCUC-CCAGCCCGUCGGCGAUGCG-UUCCGUCGGGCGAUUGGAGGGGCUGUCACGAAACAGAGUAGAGACAUU ........(((..(((((.((....)).)))))..((((((-(..(((((.(((.......-..))).)))))....)))))))((((......))))....)))..... ( -37.20, z-score = -0.02, R) >droWil1.scaffold_180955 2699636 102 + 2875958 AGGGCUUCCUCCGCCGGUGUGACGAAACAUAGGCCCCCCUC-CUAGUCCAUCGGCUACACG-UUCCGUUGGACGAUUGGAGGGGCUAAAAUGAUGAAACAAACA------ ...(.(((.((.(((.((((......)))).))).((((((-(..(((((.(((.......-..))).)))))....))))))).......)).))).).....------ ( -35.80, z-score = -1.43, R) >droVir3.scaffold_13049 20740883 99 + 25233164 AGGGCUUCCUCCGCCUGUGUGACGAAACAUCGGCCCCCCUC-CAAGCCCGUCGGCGCCGCG-CUCCGACGAACGAUUGGAGUGGCUGUUCGGUAAGUAGAA--------- ...((((((...(((.((((......)))).)))..(((((-(((...((((((.((...)-).)))))).....)))))).))......)).))))....--------- ( -35.10, z-score = -0.15, R) >droMoj3.scaffold_6680 8450302 100 + 24764193 AGGACUUCCUCCGCCUGUGUGACGAAACAUCGGCCCCCCUC-CAAGACCGUCAGCGACGCG-UUCCGUCGGACGGUUGGAGGGGCUGUUGUGAGAGUAAAUA-------- ...(((..(((((((.((((......)))).))).((((((-((..((((((..(((((..-...))))))))))))))))))).....).)))))).....-------- ( -42.00, z-score = -2.26, R) >droGri2.scaffold_15110 12775488 104 - 24565398 AGGGCUUCCUCCGCCUGUGUGACGAAACAUAGGCCCCCCUC-CAAGGCCGUCAGCGACGCG-UUCCGUCGGACGAUUGGAGGGGCUGCAAAUUUUUUAUAGCAAAC---- ...(((......((((((((......)))))))).((((((-(((...((((..(((((..-...))))))))).)))))))))...............)))....---- ( -42.80, z-score = -2.78, R) >anoGam1.chr2L 40610505 99 - 48795086 AAUGCCACCGUUGCCUGCGCCAAUGAGCAUUCGUCCCUCGGGCAUGGCAGGCAGCAGCGAGGCGGUACCGUGCGCCUGAGACUGC-GCACUCACACCGCC---------- ..(((....(((((((((((......))....((((...))))...))))))))).))).((((((...((((((........))-))))....))))))---------- ( -43.40, z-score = -1.35, R) >consensus AGGGCUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUC_CGAGACCGUCCGCAACGCG_UUCCGUCGGGCGGUUGGAGGGGCUGUCAAGAUAGAAAAGAU_______ ..(((((((((((((.((((......)))).)))..............((((..(((((......)))))))))...)))))))..)))..................... (-22.39 = -22.11 + -0.29)



| Location | 7,923,103 – 7,923,204 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Shannon entropy | 0.60041 |
| G+C content | 0.62711 |
| Mean single sequence MFE | -43.61 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.05 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7923103 101 - 24543557 -------AACCUUUCUAUAUGGACAGCCCCUCCAACCGCCCAACGGAA-CGCGUUGCGGACGGUCUCG-GAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGCCCU -------...((((((..........(((((((.(((((((((((...-..))))).)).))))...)-)))))).((((.(((......))).))))..)))))).... ( -42.80, z-score = -1.32, R) >droSim1.chr3L 7396629 102 - 22553184 ------AACCUUUCACCCCCUGACAGCCCCUCCAACCGCCCAACGGAA-CGCGUUGCGGACGGUCUCG-GAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGCCCU ------...(((((.((.........(((((((.(((((((((((...-..))))).)).))))...)-)))))).((((.(((......))).)))))).))))).... ( -44.10, z-score = -1.33, R) >droSec1.super_0 217148 102 - 21120651 ------AACCUUUCACCCCCUGACAGCCCCUCCAACCGCCCAACGGAA-CGCGUUGCGGACGGUCUCG-GAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGCCCU ------...(((((.((.........(((((((.(((((((((((...-..))))).)).))))...)-)))))).((((.(((......))).)))))).))))).... ( -44.10, z-score = -1.33, R) >droYak2.chr3L 8511048 102 - 24197627 ------AACCGCUUCUAUAUGGACAGCCCCUCCAACCGCCCAACGGAA-CGCGUUGCGGACGGUCUCG-GAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGUCCU ------..............((((..(((((((.(((((((((((...-..))))).)).))))...)-)))))).((((.(((......))).)))).......)))). ( -44.60, z-score = -1.57, R) >droEre2.scaffold_4784 22325353 101 + 25762168 ------UACCGC-UCUAUAUGAACAGCUCCUCCAGCCGCCCAACGGAA-CGCGUUGCGGACGGUCUCG-GAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGUCCU ------..((..-.............(((((((((((((((((((...-..))))).)).))).)).)-)))))).((((.(((......))).))))))(((....))) ( -40.50, z-score = -0.74, R) >droAna3.scaffold_13337 7197046 98 + 23293914 ----------AACUUCUUCCAAACAGCCCCUCCAACCGCCCGACGGAA-CGCGUCGCGGACGGUCUCG-GAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGCCCU ----------..((((((((......(((((((.(((((((((((...-..))))).)).))))...)-)))))).((((.(((......))).)))))))))))).... ( -52.10, z-score = -3.81, R) >dp4.chrXR_group8 7645113 108 + 9212921 AAUGCCUCUUCUCUAUUUCGUGACAGCCCCUCCAAUCGCCCGACGGAA-CGCAUCGCCGACGGGCUGG-GAGGGGGGCCGAUGUUUCGUAAUACCGGGGGAGGAAGUCCU ....((((((((.((((.((.(((((((((.((..(((((((.(((..-.......))).))))).))-..)))))))...)))).)).))))..))))))))....... ( -44.30, z-score = -0.72, R) >droPer1.super_24 1485873 108 - 1556852 AAUGUCUCUACUCUGUUUCGUGACAGCCCCUCCAAUCGCCCGACGGAA-CGCAUCGCCGACGGGCUGG-GAGGGGGGCCGAUGUUUCGUAAUACCGGGGGAGGAAGUCCU ....(((((...(((((....)))))(((((((....(((((.(((..-.......))).)))))..)-))))))..(((.((((....)))).))).)))))....... ( -41.70, z-score = -0.03, R) >droWil1.scaffold_180955 2699636 102 - 2875958 ------UGUUUGUUUCAUCAUUUUAGCCCCUCCAAUCGUCCAACGGAA-CGUGUAGCCGAUGGACUAG-GAGGGGGGCCUAUGUUUCGUCACACCGGCGGAGGAAGCCCU ------.((((.((((..........(((((((....(((((.(((..-.......))).)))))..)-)))))).(((..(((......)))..))))))).))))... ( -37.30, z-score = -1.43, R) >droVir3.scaffold_13049 20740883 99 - 25233164 ---------UUCUACUUACCGAACAGCCACUCCAAUCGUUCGUCGGAG-CGCGGCGCCGACGGGCUUG-GAGGGGGGCCGAUGUUUCGUCACACAGGCGGAGGAAGCCCU ---------.................((.((((((..(((((((((.(-(...)).))))))))))))-)))))((((...(.(((((((.....))))))).).)))). ( -40.70, z-score = -1.10, R) >droMoj3.scaffold_6680 8450302 100 - 24764193 --------UAUUUACUCUCACAACAGCCCCUCCAACCGUCCGACGGAA-CGCGUCGCUGACGGUCUUG-GAGGGGGGCCGAUGUUUCGUCACACAGGCGGAGGAAGUCCU --------......((((........(((((((((((((((((((...-..)))))..))))))..))-)))))).(((..(((......)))..)))))))........ ( -42.70, z-score = -2.56, R) >droGri2.scaffold_15110 12775488 104 + 24565398 ----GUUUGCUAUAAAAAAUUUGCAGCCCCUCCAAUCGUCCGACGGAA-CGCGUCGCUGACGGCCUUG-GAGGGGGGCCUAUGUUUCGUCACACAGGCGGAGGAAGCCCU ----..((((............)))).((((((((((((((((((...-..)))))..)))))..)))-)))))((((...(.(((((((.....))))))).).)))). ( -41.60, z-score = -1.96, R) >anoGam1.chr2L 40610505 99 + 48795086 ----------GGCGGUGUGAGUGC-GCAGUCUCAGGCGCACGGUACCGCCUCGCUGCUGCCUGCCAUGCCCGAGGGACGAAUGCUCAUUGGCGCAGGCAACGGUGGCAUU ----------(((((((...((((-((........))))))..)))))))..((..(((..((((.((((((((((.......))).)))).))))))).)))..))... ( -50.40, z-score = -1.82, R) >consensus _______ACCUUUUCUAUCCUGACAGCCCCUCCAACCGCCCGACGGAA_CGCGUCGCGGACGGUCUCG_GAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGCCCU .........................(((((((...(((..(((((......)))))....)))........))))))).................(((.......))).. (-17.50 = -17.05 + -0.46)

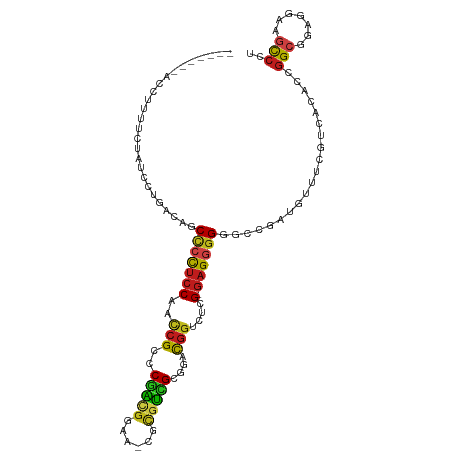
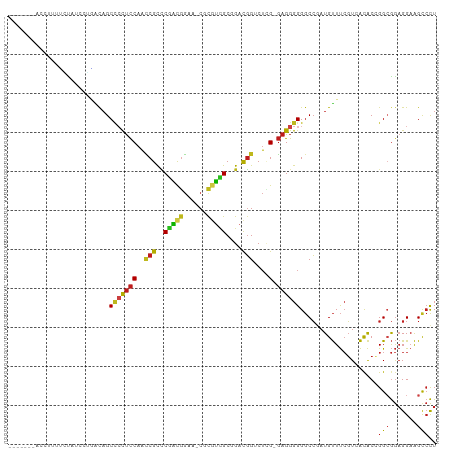
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:25 2011