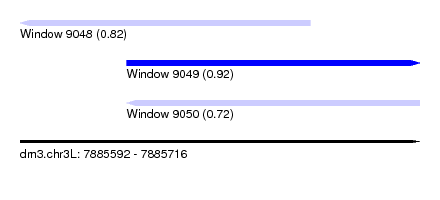
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,885,592 – 7,885,716 |
| Length | 124 |
| Max. P | 0.917168 |

| Location | 7,885,592 – 7,885,682 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 83.73 |
| Shannon entropy | 0.25364 |
| G+C content | 0.40898 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.817725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7885592 90 - 24543557 GGCCAUUUGAAAUUUGGCCAAAACGGGUGGGCAUGCAGUUUAUAUGUACAUAUAUAUGUGUGUAUAUCUGCUGGUUUUU-------UGGGGAGGCUU ((((.(((.(((...(((((...((((((.(((((((...(((((....)))))..))))))).)))))).))))).))-------).))).)))). ( -27.30, z-score = -2.00, R) >droSim1.chr3L 7368535 82 - 22553184 GGCCAUUUGAAAUUUGGCCAAAACGGGUGGGCAUGCAGUUUAUAUG------UAUAUAUGUGUAUAUCUGCUGGUUUUU---------GGGGGGCUU (((((.........)))))(((((.((..(..(((((...((((..------..))))..)))))..)..)).))))).---------......... ( -23.50, z-score = -1.29, R) >droSec1.super_0 189158 82 - 21120651 GGCCAUUUGAAAUUUGGCCAAAACGGGUGGGCAUGCAGUUUAUAUG------UAUAUAUGUGUAUAUCUGCUGGUUUUU---------GGGGGGCUU (((((.........)))))(((((.((..(..(((((...((((..------..))))..)))))..)..)).))))).---------......... ( -23.50, z-score = -1.29, R) >droEre2.scaffold_4784 22296793 97 + 25762168 GGCCAUUUGAAAAUUGGCCAAAACGGGUGGGCAUGCAGUAUACGAGCAUAUUUAUAUAUGUAUAUAUCUGCUGGCUUUUUUUUUUUUUUGGGAGCUU (((((.........)))))((((..((..(((..((((((((...((((((....)))))).)))).))))..)))..))..))))........... ( -23.30, z-score = -0.79, R) >consensus GGCCAUUUGAAAUUUGGCCAAAACGGGUGGGCAUGCAGUUUAUAUG______UAUAUAUGUGUAUAUCUGCUGGUUUUU_________GGGGGGCUU (((((.........)))))(((((.((..(..(((((...((((((......))))))..)))))..)..)).)))))................... (-21.70 = -22.32 + 0.62)

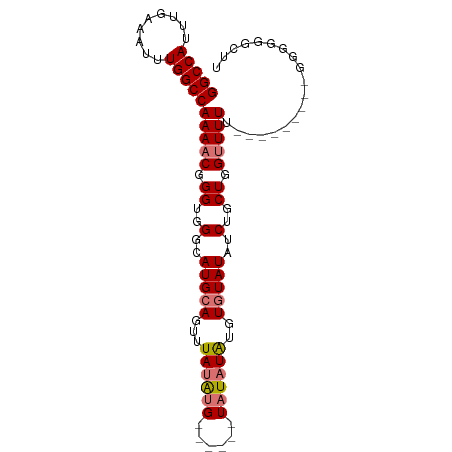
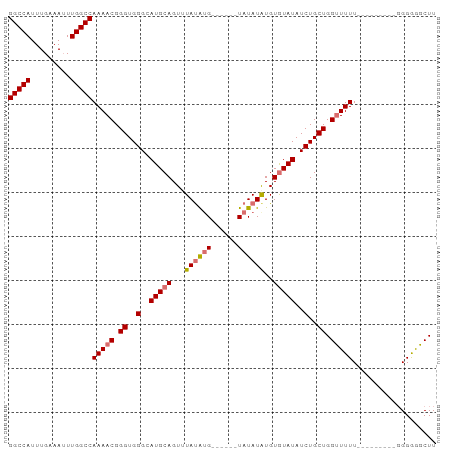
| Location | 7,885,625 – 7,885,716 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 73.71 |
| Shannon entropy | 0.45614 |
| G+C content | 0.49209 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7885625 91 + 24543557 --AUAUAUAUGUACAUAUAAACUGCAUGCCCACCCGUUUUGGCCAAAUUUCAAAUGGCCCGGGAGGUGGGCGUG-UGCCUGCCUGCCAGGU-GAU --...(((((....)))))....(((((((((((..((..(((((.........)))))..)).))))))))))-)(((((.....)))))-... ( -35.70, z-score = -2.05, R) >droSim1.chr3L 7368564 83 + 22553184 ------AUAUAUACAUAUAAACUGCAUGCCCACCCGUUUUGGCCAAAUUUCAAAUGGCCUGGGAGGUGGGCGUG-UGCCUGCC----AGGU-GAU ------.................((((((((((((.(...(((((.........))))).).).))))))))))-)(((....----.)))-... ( -32.20, z-score = -1.98, R) >droSec1.super_0 189187 83 + 21120651 ------AUAUAUACAUAUAAACUGCAUGCCCACCCGUUUUGGCCAAAUUUCAAAUGGCCUGGGAGGUGGGCGUG-UGCCUGCC----AGGU-GAU ------.................((((((((((((.(...(((((.........))))).).).))))))))))-)(((....----.)))-... ( -32.20, z-score = -1.98, R) >droEre2.scaffold_4784 22296831 89 - 25762168 AUAUAUAAAUAUGCUCGUAUACUGCAUGCCCACCCGUUUUGGCCAAUUUUCAAAUGGCCUGGGAGGUGGGCGUG-UGUCUGCC----AGGU-GAU .............((.(((.((.((((((((((((.(...(((((.........))))).).).))))))))))-))).))).----))..-... ( -33.60, z-score = -2.24, R) >droWil1.scaffold_181009 39113 93 - 3585778 -AUUUUUCGCCAGCUUGCCAACUCUGUGUGAACCUUUUUUGGCCAAUUUUCAAAUUGU-UGUCGCCAAGGCGUGGCCCCCGCCCUCUAUGUUGAU -....((((((((..(....)..))).)))))......(((((((((.........))-))..)))))((((.(....)))))............ ( -18.40, z-score = 0.13, R) >consensus ___U_UAUAUAUACAUAUAAACUGCAUGCCCACCCGUUUUGGCCAAAUUUCAAAUGGCCUGGGAGGUGGGCGUG_UGCCUGCC____AGGU_GAU ........................((((((((((......(((((.........))))).....))))))))))..................... (-19.12 = -19.88 + 0.76)
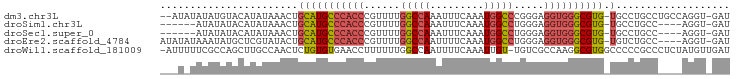

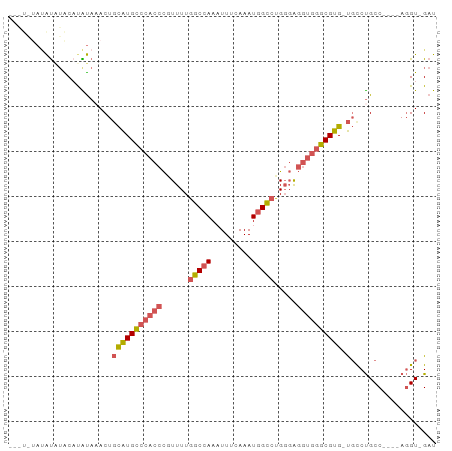
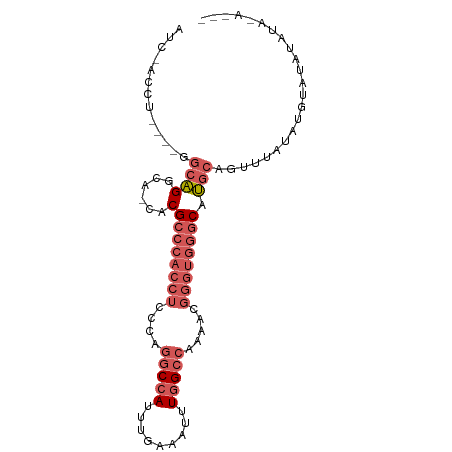
| Location | 7,885,625 – 7,885,716 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.71 |
| Shannon entropy | 0.45614 |
| G+C content | 0.49209 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -17.18 |
| Energy contribution | -19.26 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.722455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7885625 91 - 24543557 AUC-ACCUGGCAGGCAGGCA-CACGCCCACCUCCCGGGCCAUUUGAAAUUUGGCCAAAACGGGUGGGCAUGCAGUUUAUAUGUACAUAUAUAU-- ...-.((((.....))))..-...((((((((....(((((.........))))).....)))))))).((((.......)))).........-- ( -31.30, z-score = -1.32, R) >droSim1.chr3L 7368564 83 - 22553184 AUC-ACCU----GGCAGGCA-CACGCCCACCUCCCAGGCCAUUUGAAAUUUGGCCAAAACGGGUGGGCAUGCAGUUUAUAUGUAUAUAU------ ...-.((.----....))..-...((((((((....(((((.........))))).....))))))))(((((.......)))))....------ ( -27.60, z-score = -1.26, R) >droSec1.super_0 189187 83 - 21120651 AUC-ACCU----GGCAGGCA-CACGCCCACCUCCCAGGCCAUUUGAAAUUUGGCCAAAACGGGUGGGCAUGCAGUUUAUAUGUAUAUAU------ ...-.((.----....))..-...((((((((....(((((.........))))).....))))))))(((((.......)))))....------ ( -27.60, z-score = -1.26, R) >droEre2.scaffold_4784 22296831 89 + 25762168 AUC-ACCU----GGCAGACA-CACGCCCACCUCCCAGGCCAUUUGAAAAUUGGCCAAAACGGGUGGGCAUGCAGUAUACGAGCAUAUUUAUAUAU ...-....----........-...((((((((....(((((.........))))).....))))))))((((.(....)..)))).......... ( -27.30, z-score = -1.94, R) >droWil1.scaffold_181009 39113 93 + 3585778 AUCAACAUAGAGGGCGGGGGCCACGCCUUGGCGACA-ACAAUUUGAAAAUUGGCCAAAAAAGGUUCACACAGAGUUGGCAAGCUGGCGAAAAAU- ............(((....))).((((..(((....-.(((((....)))))(((((.....((....))....)))))..)))))))......- ( -22.80, z-score = -0.36, R) >consensus AUC_ACCU____GGCAGGCA_CACGCCCACCUCCCAGGCCAUUUGAAAUUUGGCCAAAACGGGUGGGCAUGCAGUUUAUAUGUAUAUAUA_A___ .............((((......)((((((((....(((((.........))))).....)))))))).)))....................... (-17.18 = -19.26 + 2.08)

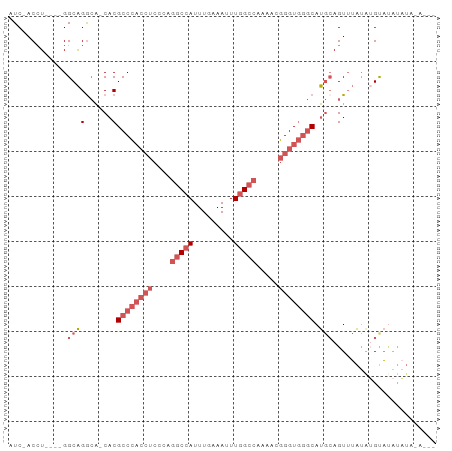
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:23 2011