
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,878,596 – 7,878,730 |
| Length | 134 |
| Max. P | 0.883551 |

| Location | 7,878,596 – 7,878,730 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 64.94 |
| Shannon entropy | 0.65960 |
| G+C content | 0.42513 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -12.56 |
| Energy contribution | -14.12 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
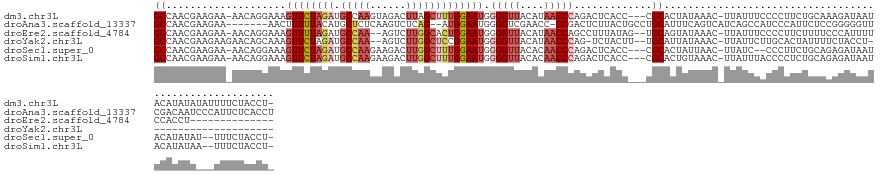
>dm3.chr3L 7878596 134 + 24543557 GCCAACGAAGAA-AACAGGAAAGUUCUAGAUGCCAAGUAGACUUAGCUUUGGAAUGGGGUUACAUAACCCAGACUCACC---CGCACUAUAAAC-UUAUUUCCCCUUCUGCAAAGAUAAUACAUAUAUAUUUUCUACCU- ........((((-((..(((((((((((((.((.(((....))).)))))))))).(((((....))))).........---............-...)))))...(((....))).............))))))....- ( -24.90, z-score = -0.81, R) >droAna3.scaffold_13337 10676210 130 + 23293914 GCCAACGAAGAA-------AACUUUUUACAUGCUCUCAAGUCUCAG--AUGGAAUGGGGUCGAACC-CCGACUCUUACUGCCUGCAUUUCAGUCAUCAGCCAUCCCAUUCUCCGGGGGUUCGACAAUCCCAUUCUCACCU ((....(((((.-------...)))))....))...........((--.(((((((((((((((((-((((((.....((....))....))))....(......)........))))))))))...))))))).)).)) ( -32.90, z-score = -1.02, R) >droEre2.scaffold_4784 22292279 120 - 25762168 GCCAACGAAGAA-AACAGGAAAGUUUUAGAUGCCAA--AGUCUUGGCACUGGAAUGGGGUUACAUAACCAGCCUUUAUAG--UGCAGUAUAAAC-UUAUUUCCCCUUCUUUUCCCAUUUUCCACCU-------------- .........(((-((..((((((((((((.((((((--....)))))))))))))((((...........((((....))--.))(((....))-).....))))....)))))..))))).....-------------- ( -29.00, z-score = -1.51, R) >droYak2.chr3L 8477570 113 + 24197627 GCCAACGAAGAAGAACAGCAAAGUUCUAGAUGCCAA--AGUCUUGGCUCCGGAAUGGGGUUACAUAACCCAG-UCUACUU--UGCAUUAUAAAC-UUAUUCUUGCACUAUUUUCUACCU--------------------- ........((((((...(((((((((..((.(((((--....)))))))..))...(((((....)))))..-...))))--)))..((.....-.))...........))))))....--------------------- ( -25.80, z-score = -1.27, R) >droSec1.super_0 184688 130 + 21120651 GCCAACGAAGAA-AACAGGAAAGUUCUAGAUGCCAAGAAGACUUGGCUUUGGAAUGGGGUUACACAACCCAGACUCACC---CGCACUAUUAAC-UUAUC--CCCUUCUGCAGAGAUAAUACAUAUAU--UUUCUACCU- ........((((-((..((...((((((((.((((((....)))))))))))))).(((((....)))))........)---)...........-(((((--.(........).)))))........)--)))))....- ( -28.30, z-score = -1.38, R) >droSim1.chr3L 7364039 132 + 22553184 GCCAACGAAGAA-AACAGGAAAGUUCUAGAUGCCAAGAAGACUUGGCUUUGGAAUGGGGUUACACAACCCAGACUCACC---CGCACUGUAAAC-UUAUUUACCCCUCUGCAGAGAUAAUACAUAUAA--UUUCUACCU- ........((((-(((((....((((((((.((((((....)))))))))))))).(((((....))))).........---....))))....-..........(((....))).............--)))))....- ( -30.80, z-score = -1.95, R) >consensus GCCAACGAAGAA_AACAGGAAAGUUCUAGAUGCCAAGAAGACUUGGCUUUGGAAUGGGGUUACACAACCCAGACUCACC___CGCACUAUAAAC_UUAUUUACCCUUCUGCACAGAUAAUACAUAUA___UUUCUACCU_ ((....................((((((((.(((((......))))))))))))).(((((....))))).............))....................................................... (-12.56 = -14.12 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:19 2011