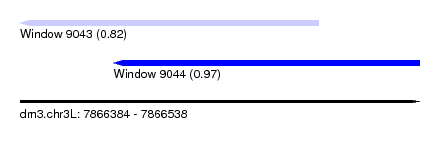
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,866,384 – 7,866,538 |
| Length | 154 |
| Max. P | 0.974900 |

| Location | 7,866,384 – 7,866,499 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.31 |
| Shannon entropy | 0.39074 |
| G+C content | 0.33438 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -15.99 |
| Energy contribution | -15.99 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
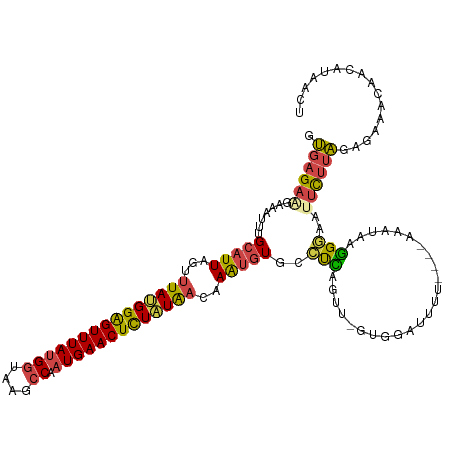
>dm3.chr3L 7866384 115 - 24543557 GUGAGAAGAAAUUUGCAUUAGUUUAUGGAGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGCCUCACUU-UUGGAUUUU----AAAUAAGGCAAUUCUUAGAGAAACAACAUUACU .((((((.......(((((.(((.(((((((((((((....)).)))))))))))))).)))))((((....(-(((.....)----)))..))))..))))))................ ( -28.10, z-score = -2.69, R) >droAna3.scaffold_13337 10660911 119 - 23293914 UCGAGUGGAUAUUUGCAUUCAACUAAGGAGUUUAAGCAAAGCCAAUGAACUUUAUGAAAAAAAUAUUCUAUCUAGAGGAGCUUGACAAGAAAAAGACAUUUAUGGACAGAUCACUUAUU- ..((((((((((((...((((.....((((((((.((...))...)))))))).))))..))))))(((.((((......(((.........))).......)))).)))))))))...- ( -16.82, z-score = 0.77, R) >droEre2.scaffold_4784 22277679 115 + 25762168 GUGAGAUGAAAUUUGCAUUAGUUUAUGGGGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGCCUUAGUU-GUGCAUUUU----AAAUAAGGGAAUUCUGAGAGAAACAACUUUGCU ..............(((((.(((.(((((((((((((....)).)))))))))))))).)))))((...((((-((...((((----((((......))).)))))...))))))..)). ( -28.00, z-score = -1.84, R) >droYak2.chr3L 8464804 115 - 24197627 GUGAGAUGAAAUUUGCAUUAGCUUACGGAGUUUAUGGCAAGCCAAUGAACUCUGUAAAAAGUGUGCCUCAGUU-GUGGAUAUU----AAAUAAGGGAAUUCUUAGAGCAACAACAUUGCU ....((((......((((....(((((((((((((((....)).))))))))))))).....))))....(((-((.......----...(((((....)))))..)))))..))))... ( -28.30, z-score = -1.24, R) >droSim1.chr3L 7352003 115 - 22553184 GUGAGAAGAAAUUUGCAUUAGUUUAUGGAGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGACUCGGUU-GUGGAUUUU----AAAAAAGGGAAUUCUUAGAGAAACAACAUAACU ..(((.......(..((((.(((.(((((((((((((....)).)))))))))))))).))))..))))((((-(((...(((----....((((....))))....)))...))))))) ( -28.01, z-score = -2.97, R) >droSec1.super_0 172760 115 - 21120651 GUGAGAAGAAAUUUGCAUUAGUUUAUGGAGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGACUCGGUU-GUGGAUUUU----AAAUAAGGGAAUUCUUAGAGAGACAACGAAACU .((((.......(..((((.(((.(((((((((((((....)).)))))))))))))).))))..)))))(((-((...((((----...(((((....))))).)))))))))...... ( -27.91, z-score = -2.70, R) >consensus GUGAGAAGAAAUUUGCAUUAGUUUAUGGAGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGCCUCAGUU_GUGGAUUUU____AAAUAAGGGAAUUCUUAGAGAAACAACAUAACU .((((((.......(((((...(((((((((((((((....)).)))))))))))))..)))))..(((........................)))..))))))................ (-15.99 = -15.99 + 0.01)

| Location | 7,866,420 – 7,866,538 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.03 |
| Shannon entropy | 0.32826 |
| G+C content | 0.35782 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -23.47 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
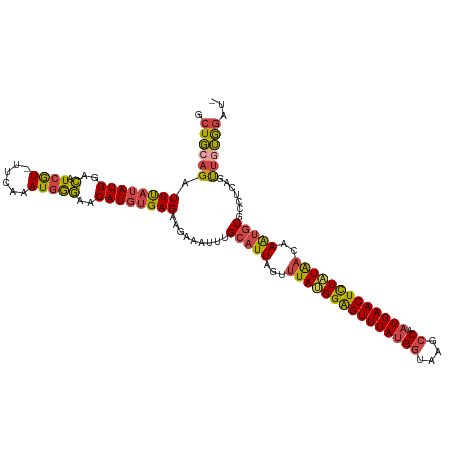
>dm3.chr3L 7866420 118 - 24543557 GCUGCAGAUUUAUAUGGACAUCGU-UUUAAAUGGGAACAUGUGAGAAGAAAUUUGCAUUAGUUUAUGGAGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGCCUCACUUUUGGAU- ..(((((((((......((((.((-(((.....)))))))))......))))))))).....(((((((((((((((....)).)))))))))))))((((.(((...))).))))...- ( -32.20, z-score = -2.82, R) >droAna3.scaffold_13337 10660950 119 - 23293914 ACUUACGAUUUAUAUGGGUAUAAU-UUUAAAUGAAUCCAUUCGAGUGGAUAUUUGCAUUCAACUAAGGAGUUUAAGCAAAGCCAAUGAACUUUAUGAAAAAAAUAUUCUAUCUAGAGGAG .(((....((((.(((((.(((.(-(((..((((((((((....)))))).(((((.((.((((....)))).)))))))...........))))...)))).)))))))).)))).))) ( -17.60, z-score = 0.35, R) >droEre2.scaffold_4784 22277715 119 + 25762168 GCUGCAGAUUUAUAUGGACAUCGUGUUGAAAUGGGAACAUGUGAGAUGAAAUUUGCAUUAGUUUAUGGGGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGCCUUAGUUGUGCAU- ..((((.(.(((...((.(.((((.((...(((....)))..)).)))).....(((((.(((.(((((((((((((....)).)))))))))))))).)))))))).))).).)))).- ( -31.80, z-score = -1.74, R) >droYak2.chr3L 8464840 118 - 24197627 GCUGCAGAUUUAUAUGGACAUCGU-UUCAAAUGGGAACAUGUGAGAUGAAAUUUGCAUUAGCUUACGGAGUUUAUGGCAAGCCAAUGAACUCUGUAAAAAGUGUGCCUCAGUUGUGGAU- .(..(((.............((((-((((.(((....))).)))))))).....((((....(((((((((((((((....)).))))))))))))).....)))).....)))..)..- ( -37.70, z-score = -3.23, R) >droSim1.chr3L 7352039 118 - 22553184 GCUGCAGAUUUAUAUGGACAUCGU-UUCAAAUGGGAACAUGUGAGAAGAAAUUUGCAUUAGUUUAUGGAGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGACUCGGUUGUGGAU- .(..(((.............((.(-((((.(((....))).))))).))...(..((((.(((.(((((((((((((....)).)))))))))))))).))))..).....)))..)..- ( -33.50, z-score = -3.06, R) >droSec1.super_0 172796 118 - 21120651 GCUGCAGAUUCAUAUGGACAUCGU-UUCAAAUGGGAACAUGUGAGAAGAAAUUUGCAUUAGUUUAUGGAGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGACUCGGUUGUGGAU- .(..(((.((((((((..(.((((-.....)))))..)))))))).......(..((((.(((.(((((((((((((....)).)))))))))))))).))))..).....)))..)..- ( -34.50, z-score = -3.05, R) >consensus GCUGCAGAUUUAUAUGGACAUCGU_UUCAAAUGGGAACAUGUGAGAAGAAAUUUGCAUUAGUUUAUGGAGUUUAUGGUAAGCCAAUGAACUCUAUAACAAAUGUGCCUCAGUUGUGGAU_ .((((((.((((((((..(.((((......)))))..)))))))).........(((((...(((((((((((((((....)).)))))))))))))..))))).......))))))... (-23.47 = -23.62 + 0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:19 2011