
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,865,084 – 7,865,158 |
| Length | 74 |
| Max. P | 0.974079 |

| Location | 7,865,084 – 7,865,158 |
|---|---|
| Length | 74 |
| Sequences | 10 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 92.48 |
| Shannon entropy | 0.15607 |
| G+C content | 0.41264 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -16.18 |
| Energy contribution | -16.04 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7865084 74 + 24543557 GCCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGCU-GCCGAUAAAGCGUAAUGAGAAAAAGUGGCA ..(((((((((...(((((..........((((((.....))))-)))))))...)))))))))........... ( -24.20, z-score = -3.06, R) >droSec1.super_0 171505 74 + 21120651 GCCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGCU-GCCGAUAAAGCGUAAUGAGAAAAAGUGGCA ..(((((((((...(((((..........((((((.....))))-)))))))...)))))))))........... ( -24.20, z-score = -3.06, R) >droSim1.chr3L 7350737 74 + 22553184 GCCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGCU-GCCGAUAAAGCGUAAUGAGAAAAAGUGGCA ..(((((((((...(((((..........((((((.....))))-)))))))...)))))))))........... ( -24.20, z-score = -3.06, R) >droYak2.chr3L 8463548 74 + 24197627 GCCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGCU-GCCGAUAAAACGUAAUGAGAAAAAGUGGCA ..(((((((((...(((((..........((((((.....))))-)))))))...)))))))))........... ( -23.10, z-score = -3.20, R) >droEre2.scaffold_4784 22276395 74 - 25762168 GCCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGCU-GCCGAUAAAACGUAAUGAGAAAAAGUGGCA ..(((((((((...(((((..........((((((.....))))-)))))))...)))))))))........... ( -23.10, z-score = -3.20, R) >droAna3.scaffold_13337 10659920 75 + 23293914 GUCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGCCCGGCGAUAAAACGUAAUGAGAAAAAGUAGCA .((((((((((...(((((((..(......)((((.....)))).)))))))...)))))))))).......... ( -28.90, z-score = -5.56, R) >dp4.chrXR_group8 7594543 74 - 9212921 GUCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGAC-GCCGAUAAAACGUAAUGAGAAAAAGUAGCG .((((((((((....(((.....)))((((((((.((....)).-)))).)))).)))))))))).......... ( -20.80, z-score = -3.00, R) >droPer1.super_24 1434060 74 + 1556852 GUCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGAC-GCCGAUAAAACGUAAUGAGAAAAAGUAGCG .((((((((((....(((.....)))((((((((.((....)).-)))).)))).)))))))))).......... ( -20.80, z-score = -3.00, R) >droWil1.scaffold_181009 8930 73 - 3585778 GUCUCAUUAUGAAAUGUCGUCAAGAUUU-GCGGCUUUAAUAGUU-GUCAAUAAAAUGUAAUGAGAAAAAGUAGCA .(((((((((.....(((.....)))((-((((((.....))))-).)))......))))))))).......... ( -14.80, z-score = -2.05, R) >droMoj3.scaffold_6680 16372321 73 - 24764193 GCCUCAUUACGAAAUGUCGCCAAGAUUU-GCGGCUUUAAUAGAC-GCCGUUAAAACGUAAUGGGGAAAAGCAGCA .((((((((((....(((.....)))..-(((((.((....)).-))))).....)))))))))).......... ( -21.90, z-score = -2.10, R) >consensus GCCUCAUUACGAAAUGUCGCCAAGAUUUUGCGGCUUUAAUAGCU_GCCGAUAAAACGUAAUGAGAAAAAGUAGCA ..(((((((((...((((((.........))(((...........)))))))...)))))))))........... (-16.18 = -16.04 + -0.14)

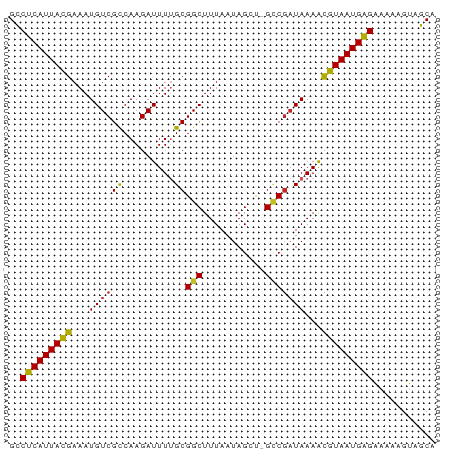
| Location | 7,865,084 – 7,865,158 |
|---|---|
| Length | 74 |
| Sequences | 10 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 92.48 |
| Shannon entropy | 0.15607 |
| G+C content | 0.41264 |
| Mean single sequence MFE | -17.94 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.71 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7865084 74 - 24543557 UGCCACUUUUUCUCAUUACGCUUUAUCGGC-AGCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGGC ..........((((((((((((......((-.(((.....))).))........)))((....))))))))))). ( -18.34, z-score = -1.68, R) >droSec1.super_0 171505 74 - 21120651 UGCCACUUUUUCUCAUUACGCUUUAUCGGC-AGCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGGC ..........((((((((((((......((-.(((.....))).))........)))((....))))))))))). ( -18.34, z-score = -1.68, R) >droSim1.chr3L 7350737 74 - 22553184 UGCCACUUUUUCUCAUUACGCUUUAUCGGC-AGCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGGC ..........((((((((((((......((-.(((.....))).))........)))((....))))))))))). ( -18.34, z-score = -1.68, R) >droYak2.chr3L 8463548 74 - 24197627 UGCCACUUUUUCUCAUUACGUUUUAUCGGC-AGCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGGC ..........((((((((((........((-.(((.....))).)).......((....))...)))))))))). ( -18.10, z-score = -1.86, R) >droEre2.scaffold_4784 22276395 74 + 25762168 UGCCACUUUUUCUCAUUACGUUUUAUCGGC-AGCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGGC ..........((((((((((........((-.(((.....))).)).......((....))...)))))))))). ( -18.10, z-score = -1.86, R) >droAna3.scaffold_13337 10659920 75 - 23293914 UGCUACUUUUUCUCAUUACGUUUUAUCGCCGGGCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGAC ..........((((((((((.....((((((((((.....)))).........)))))).....)))))))))). ( -25.90, z-score = -4.94, R) >dp4.chrXR_group8 7594543 74 + 9212921 CGCUACUUUUUCUCAUUACGUUUUAUCGGC-GUCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGAC ..........((((((((((.....(((((-..........)))((.........)))).....)))))))))). ( -19.20, z-score = -2.91, R) >droPer1.super_24 1434060 74 - 1556852 CGCUACUUUUUCUCAUUACGUUUUAUCGGC-GUCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGAC ..........((((((((((.....(((((-..........)))((.........)))).....)))))))))). ( -19.20, z-score = -2.91, R) >droWil1.scaffold_181009 8930 73 + 3585778 UGCUACUUUUUCUCAUUACAUUUUAUUGAC-AACUAUUAAAGCCGC-AAAUCUUGACGACAUUUCAUAAUGAGAC ..........((((((((((......))..-..........(.(((-(.....)).)).)......)))))))). ( -7.90, z-score = -1.24, R) >droMoj3.scaffold_6680 16372321 73 + 24764193 UGCUGCUUUUCCCCAUUACGUUUUAACGGC-GUCUAUUAAAGCCGC-AAAUCUUGGCGACAUUUCGUAAUGAGGC ...........(((((((((......((((-..........)))).-......((....))...))))))).)). ( -16.00, z-score = -0.44, R) >consensus UGCCACUUUUUCUCAUUACGUUUUAUCGGC_AGCUAUUAAAGCCGCAAAAUCUUGGCGACAUUUCGUAAUGAGGC ..........(((((((((((((((..((....))..))))))(((.........))).......))))))))). (-15.65 = -15.71 + 0.06)

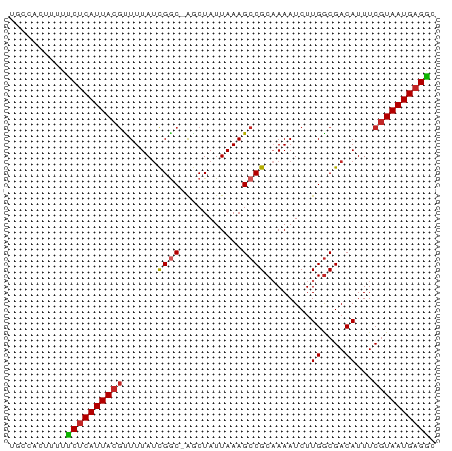
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:17 2011