| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,590,814 – 4,590,924 |
| Length | 110 |
| Max. P | 0.788575 |

| Location | 4,590,814 – 4,590,924 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.68 |
| Shannon entropy | 0.32677 |
| G+C content | 0.40475 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -17.62 |
| Energy contribution | -18.09 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
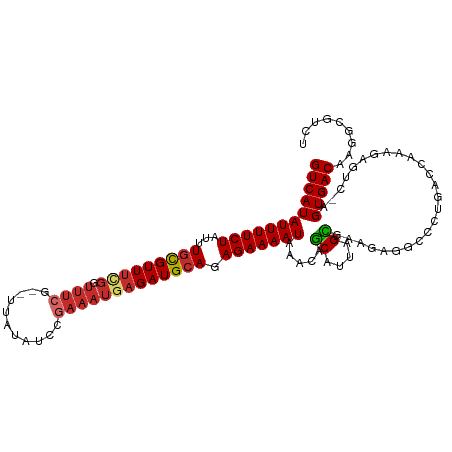
>dm3.chr2L 4590814 110 + 23011544 GUCAUAUUUUCUAUUUGCGUUUCGGUUUCG--UUACAUCCGAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGCGAAGAGGCCCUGACCAAAGAGUC-AGUGACAAGGCGUCU .((.((((((((...(((((((((.(((((--.......)))))))))))))).))))))))....((....))))(((.((((((((......)))-))......))).))) ( -34.00, z-score = -3.45, R) >droSim1.chr2L 4477319 110 + 22036055 GUCAUAUUUUCUAUUUGCGUUUCGGUUUCG--UUACAUCCGAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGCGAAGAGGCCCUGACCAAAGAGUC-AGUGACAAGGCGUCU .((.((((((((...(((((((((.(((((--.......)))))))))))))).))))))))....((....))))(((.((((((((......)))-))......))).))) ( -34.00, z-score = -3.45, R) >droSec1.super_5 2683804 110 + 5866729 GUCAUAUUUUCUAUUUGCGUUUCGGUUUCG--UUACAUCCGAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGCGAAGAGGCCCUGACCAAAGAGUC-AGUGACAAGGCGUCU .((.((((((((...(((((((((.(((((--.......)))))))))))))).))))))))....((....))))(((.((((((((......)))-))......))).))) ( -34.00, z-score = -3.45, R) >droYak2.chr2L 4611571 110 + 22324452 GUCAUAUUUUCUAUUUGCGUUUCGGUUUCG--UUAUAUCCGAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGCGAAGAGGCCCUGACCAAAGAGUC-AGUGACAAGGCGUCU .((.((((((((...(((((((((.(((((--.......)))))))))))))).))))))))....((....))))(((.((((((((......)))-))......))).))) ( -34.00, z-score = -3.59, R) >droEre2.scaffold_4929 4676821 110 + 26641161 GUCAUAUUUUCUAUUUGCGUUUCGGUUUCG--UUAUAUCCGAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGCGAAGAGGCCCUGACCACAGAGUC-AGUGACAAGCCGUCU .((.((((((((...(((((((((.(((((--.......)))))))))))))).))))))))....((....))))((((((((((((......)))-)).)....))).))) ( -31.60, z-score = -3.12, R) >droAna3.scaffold_12943 1789508 109 - 5039921 GUCAUAUUUUCUAUUUGCGUU-CGGUUUCG--UUAGUUUUGAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGUGAAGAGUCCCUGACCGAAGAGCC-GGUGACAAGGCGUCU (((.((((((((...((((((-..((((((--.......))))))..)))))).)))))))).................(((....((((......)-))))))..))).... ( -26.70, z-score = -0.94, R) >dp4.chr4_group3 438833 110 - 11692001 GUCAUAUUUUCUAUUUGUGUUUGGGUUUCG--UC-UUCCACAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGUGGAGAGCUCAUGACCAAAAAUCCCAGUGACAAGGCGUCU (((((........((((.((((((((((..--..-.(((((.((((...((.............)).)))).)))))))))))).))))))).......)))))......... ( -21.98, z-score = 0.39, R) >droPer1.super_8 1478096 110 - 3966273 GUCAUAUUUUCUAUUUGUGUUUGGGUUUCG--UC-UUCCACAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGUGGAGAGCUCAUGACCAAAAAUCCCAGUGACAAGGCGUCU (((((........((((.((((((((((..--..-.(((((.((((...((.............)).)))).)))))))))))).))))))).......)))))......... ( -21.98, z-score = 0.39, R) >droWil1.scaffold_180703 1332652 92 - 3946847 GUCAUAUUUUCUAUUUAUGUUUUUUCAACAAAUUUUUGUCGAAAUGAGAUGCAGAGAAAAUAAACAACAUUAGU---------------------UUGAGUGACAAGGCGUCU (((((.((((((...(((.((.((((.((((....)))).)))).)).)))...))))))(((((.......))---------------------))).)))))......... ( -17.30, z-score = -0.87, R) >consensus GUCAUAUUUUCUAUUUGCGUUUCGGUUUCG__UUAUAUCCGAAAUGAGAUGCAGAGAAAAUAAACAGCAUUAGCGAAGAGGCCCUGACCAAAGAGUC_AGUGACAAGGCGUCU ((((((((((((...(((((((((.((((...........))))))))))))).))))))).....((....)).........................)))))......... (-17.62 = -18.09 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:47 2011