| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,810,966 – 7,811,074 |
| Length | 108 |
| Max. P | 0.999859 |

| Location | 7,810,966 – 7,811,074 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.19 |
| Shannon entropy | 0.39940 |
| G+C content | 0.32080 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -15.49 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
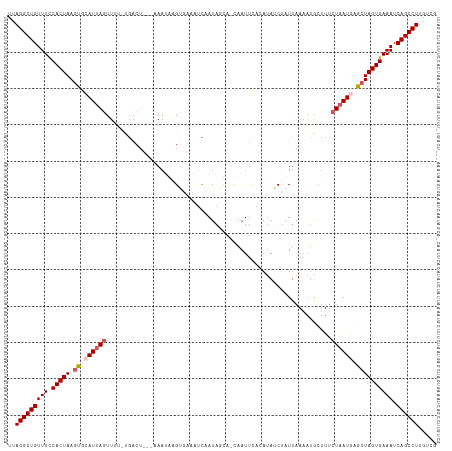
>dm3.chr3L 7810966 108 + 24543557 UUAGGCUGUUUUCACUAAGUGCAUAAGUUUU-UGACU---GAAUUCGU-----CAAUAGGA-CAAUUCACACAUCUAUUAAACGGAUUUCUAAUGAACUAGUGAAAUCAGCCUUGUCG ..((((((.((((((((.((.(((.((....-....(---(((((.((-----(.....))-)))))))...((((.......))))..)).))).)))))))))).))))))..... ( -28.90, z-score = -3.44, R) >droSim1.chr3L 7298334 113 + 22553184 UUAGGCUGUUUCCACUAAGUGCAUUAGUUUU-UGACU---CAAAAAGUGAAUUCAAUAGUA-UAAUUCACAUAACUAUUAAAAUACUUUCUAAUGCGCUAGUGAAAUCAGCCUUGUCG ..((((((((((.((((.(((((((((((((-((...---))))))(((((((........-.)))))))...................))))))))))))))))).))))))..... ( -34.10, z-score = -5.92, R) >droSec1.super_0 117434 113 + 21120651 UUAGGCUGUUUCCACUAAGUGCAUUAGUUUU-UGACU---CAAAAAGUGAAUUCAAUAGUA-UAAUUCACAUAACUAUUAAAAUACUUUCUAAUGAGCUAGUGAAAUCAGCCUUGUCG ..((((((((((.((((.((.((((((((((-((...---))))))(((((((........-.)))))))...................)))))).)))))))))).))))))..... ( -28.20, z-score = -3.71, R) >droYak2.chr3L 8407460 107 + 24197627 UUAGGCUGUUUCCACUAAGUGCAUUAGUU-----------AAAUGUGAUAAAUUUAUAUCCGCAAUUUACAUAUCGUUUUAAAUGCUUUCUAAUAAACUAGUGAAAUCAGCCUUUUCG ..((((((((((.((((.((..(((((..-----------..((((((.....))))))..(((.((.((.....))...)).)))...)))))..)))))))))).))))))..... ( -22.40, z-score = -2.99, R) >droEre2.scaffold_4784 22221798 117 - 25762168 UUAGGCUGUUUCCACUAACUGCAUUAUUUUAACGACUCAGAAAUGGGAUACAUCUAUAUCAGCAAUUUACAUAUCAUUUAAUAUGCUUUCUAAUAAGCUAGUGAAAUCAGCCUUUCG- ..((((((((((.((((..(((..(((........((((....))))........)))...)))....................((((......)))))))))))).))))))....- ( -24.29, z-score = -2.73, R) >consensus UUAGGCUGUUUCCACUAAGUGCAUUAGUUUU_UGACU___AAAUAAGUGAAAUCAAUAGCA_CAAUUCACAUAUCUAUUAAAAUGCUUUCUAAUGAGCUAGUGAAAUCAGCCUUGUCG ..(((((((((.(((((.((.((((((..............................................................)))))).)))))))))).))))))..... (-15.49 = -16.25 + 0.76)


| Location | 7,810,966 – 7,811,074 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Shannon entropy | 0.39940 |
| G+C content | 0.32080 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -19.83 |
| Energy contribution | -19.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -5.01 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.60 |
| SVM RNA-class probability | 0.999859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
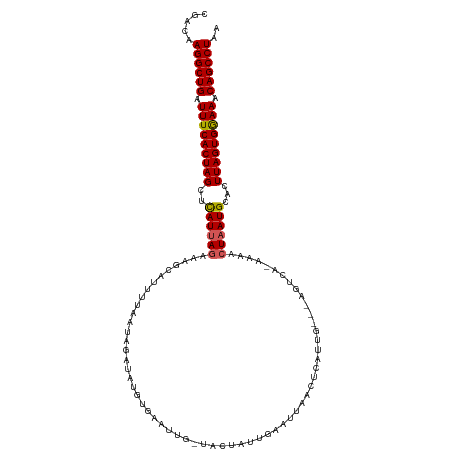
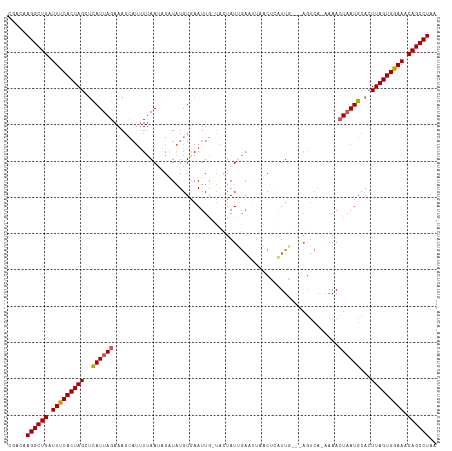
>dm3.chr3L 7810966 108 - 24543557 CGACAAGGCUGAUUUCACUAGUUCAUUAGAAAUCCGUUUAAUAGAUGUGUGAAUUG-UCCUAUUG-----ACGAAUUC---AGUCA-AAAACUUAUGCACUUAGUGAAAACAGCCUAA .....((((((.((((((((((.(((.((.....((((.....))))..(((((((-((.....)-----)).)))))---)....-....)).))).)).)))))))).)))))).. ( -30.20, z-score = -3.97, R) >droSim1.chr3L 7298334 113 - 22553184 CGACAAGGCUGAUUUCACUAGCGCAUUAGAAAGUAUUUUAAUAGUUAUGUGAAUUA-UACUAUUGAAUUCACUUUUUG---AGUCA-AAAACUAAUGCACUUAGUGGAAACAGCCUAA .....((((((.(((((((((.((((((((((((..(((((((((.(((.....))-))))))))))...)))))(((---...))-)...))))))).).)))))))).)))))).. ( -37.20, z-score = -6.05, R) >droSec1.super_0 117434 113 - 21120651 CGACAAGGCUGAUUUCACUAGCUCAUUAGAAAGUAUUUUAAUAGUUAUGUGAAUUA-UACUAUUGAAUUCACUUUUUG---AGUCA-AAAACUAAUGCACUUAGUGGAAACAGCCUAA .....((((((.(((((((((..(((((((((((..(((((((((.(((.....))-))))))))))...)))))(((---...))-)...))))))...))))))))).)))))).. ( -32.70, z-score = -4.61, R) >droYak2.chr3L 8407460 107 - 24197627 CGAAAAGGCUGAUUUCACUAGUUUAUUAGAAAGCAUUUAAAACGAUAUGUAAAUUGCGGAUAUAAAUUUAUCACAUUU-----------AACUAAUGCACUUAGUGGAAACAGCCUAA .....((((((.((((((((((.((((((...(((((((..........)))).))).((((......))))......-----------..)))))).)).)))))))).)))))).. ( -29.80, z-score = -5.23, R) >droEre2.scaffold_4784 22221798 117 + 25762168 -CGAAAGGCUGAUUUCACUAGCUUAUUAGAAAGCAUAUUAAAUGAUAUGUAAAUUGCUGAUAUAGAUGUAUCCCAUUUCUGAGUCGUUAAAAUAAUGCAGUUAGUGGAAACAGCCUAA -....((((((.(((((((((((..((((((((((((((....)))))))........(((((....)))))...)))))))(.(((((...))))))))))))))))).)))))).. ( -36.50, z-score = -5.20, R) >consensus CGACAAGGCUGAUUUCACUAGCUCAUUAGAAAGCAUUUUAAUAGAUAUGUGAAUUG_UACUAUUGAAUUAACUCAUUG___AGUCA_AAAACUAAUGCACUUAGUGGAAACAGCCUAA .....((((((.(((((((((..((((((..............................................................))))))...))))))))).)))))).. (-19.83 = -19.83 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:12 2011