| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,784,085 – 7,784,236 |
| Length | 151 |
| Max. P | 0.920361 |

| Location | 7,784,085 – 7,784,196 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Shannon entropy | 0.41970 |
| G+C content | 0.44632 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -17.46 |
| Energy contribution | -16.69 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
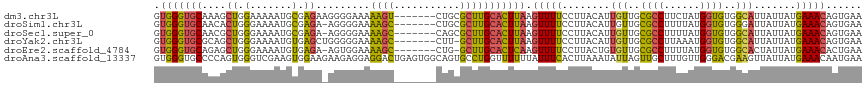
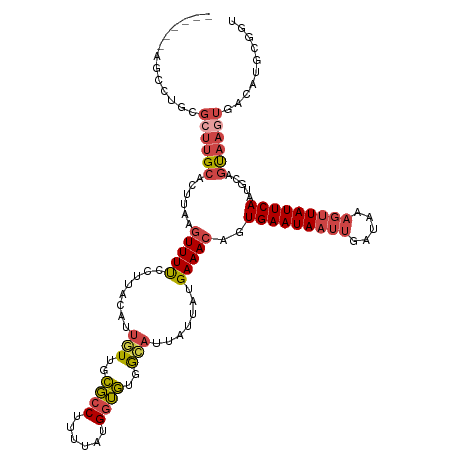
>dm3.chr3L 7784085 111 + 24543557 GUGGGUGCAAAGCUGGAAAAAUGCGAGAAGGGGAAAAAGU-------CUGCGCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUCUAUGGUGUGGCAUUAUUAUGAAACAGUGAA (((((.(.((((((....((.((((((..(.(((.....)-------)).).)))))).)).))))))))))))..(((..((((......))))..))).................. ( -29.60, z-score = -0.49, R) >droSim1.chr3L 7278555 110 + 22553184 GUGGGUGCAACACUGGGAAAAUGCGAGA-AGGGGAAAAGC-------CUGCGCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUUUAUGGUGUGGGAUUAUUAUGAAACAGUGAA ..((((((((((..(((((((((((((.-.(((......)-------))...))))))......))))))).....))))))))))(((((((((......)))))))))........ ( -35.80, z-score = -2.45, R) >droSec1.super_0 97878 110 + 21120651 GUGGGUGCAACGCUGGGAAAAUGCGAGA-AGGGGAAAAGC-------CAGCGCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUUUAUGGUGUGGCAUUAUUAUGAAACAGUGAA .(((((((((((((((.....(.(....-.).)......)-------))))).))))))))).(((((........(((..((((......))))..))).......)))))...... ( -36.76, z-score = -2.10, R) >droYak2.chr3L 8385746 110 + 24197627 GUGGGUGCGCAGCUGGGAAAAUGUGAGCUGGGGGAAAAGC-------CUU-GCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUAAAUGGUGUGGCAUUAUUAUGAAACAGUGAA .((((.(((((((.(((((((((..(((.(((.......)-------)).-)))..))......)))))))......)))))))))))...(.(((..(((....)))..))).)... ( -34.20, z-score = -1.18, R) >droEre2.scaffold_4784 22201704 109 - 25762168 GUGGGUGCAGAGCUGGGAAAAUGUGAGA-AGUGGAAAAGC-------CUG-GCUUGCACUCAAGUUUUCCUUACUGUGUUGCGCCUUUUAUGGUGUGGCACUAUUAUGAAACACUGAA .((((((((.(((..((...........-..........)-------)..-)))))))))))(((((((......((((..((((......))))..))))......)))).)))... ( -38.70, z-score = -3.02, R) >droAna3.scaffold_13337 10583795 118 + 23293914 GUGGGUGCCCCAGUGGGUCGAAGUGGAAGAAGAGGAGGACUGAGUGGCAGUGCCUGGUUUUUUAUUUCACUUAAAUAUUAGUUGCUUUGUUGGGACGAAGUUAUUAUGAAACAAUGAA .((((...)))).((..(((((((((((((..(((...((((.....)))).)))..)))))))))))..........((((.(((((((....))))))).)))).))..))..... ( -31.10, z-score = -1.04, R) >consensus GUGGGUGCAAAGCUGGGAAAAUGCGAGA_AGGGGAAAAGC_______CUGCGCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUUUAUGGUGUGGCAUUAUUAUGAAACAGUGAA .(((((((....((.(.......).)).........((((...........))))))))))).(((((........(((..((((......))))..))).......)))))...... (-17.46 = -16.69 + -0.77)
| Location | 7,784,122 – 7,784,236 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Shannon entropy | 0.38493 |
| G+C content | 0.37026 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.84 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
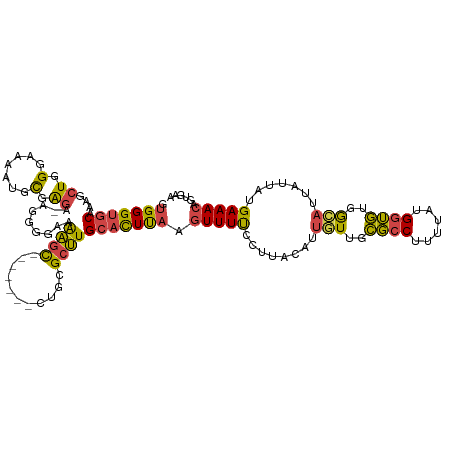
>dm3.chr3L 7784122 114 + 24543557 ------AGUCUGCGCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUCUAUGGUGUGGCAUUAUUAUGAAACAGUGAAUAAUUGAUAAAGUUAUUCAAUGCAGUAAGUGACAUGCGGU ------......(((.((((((((.(((((........(((..((((......))))..))).......)))))..(((((((((.....)))))))))......)))))).)).))).. ( -34.76, z-score = -2.74, R) >droSim1.chr3L 7278591 114 + 22553184 ------AGCCUGCGCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUUUAUGGUGUGGGAUUAUUAUGAAACAGUGAAUAAUUGAUAAAGUUAUUCAAUGCAGCAAGUGACAUGCGGU ------.((.(((((((((......(((((........(.(..((((......))))..).).......)))))..(((((((((.....))))))))).....))))))).)).))... ( -33.76, z-score = -2.14, R) >droSec1.super_0 97914 114 + 21120651 ------AGCCAGCGCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUUUAUGGUGUGGCAUUAUUAUGAAACAGUGAAUAAUUGAUAAAGUUAUUCAAUGCAGUAAGUGACAUGCGGU ------......(((.((((((((.(((((........(((..((((......))))..))).......)))))..(((((((((.....)))))))))......)))))).)).))).. ( -34.76, z-score = -2.58, R) >droYak2.chr3L 8385783 113 + 24197627 ------AGCCUU-GCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUAAAUGGUGUGGCAUUAUUAUGAAACAGUGAAUAAUUGAAAAACUUAUUCAAUGCCGUAAGUGACAUGCGGU ------.(((..-((.((((((((.(((((........(((..((((......))))..))).......)))))..(((((((((....)).)))))))......)))))).)).))))) ( -31.36, z-score = -2.44, R) >droEre2.scaffold_4784 22201740 113 - 25762168 ------AGCCUG-GCUUGCACUCAAGUUUUCCUUACUGUGUUGCGCCUUUUAUGGUGUGGCACUAUUAUGAAACACUGAAUAAUUGAUAAAGCUAUUCAAUGCAGUAAUUCACAUGCGGU ------.((.((-(((((....)))))..........((((..((((......))))..)))).....((((..((((....(((((.........))))).))))..)))))).))... ( -30.00, z-score = -1.31, R) >droAna3.scaffold_13337 10583833 106 + 23293914 ACUGAGUGGCAGUGCCUGGUUUUUUAUUUCACUUAAAUAUUAGUUGCUUUGUUGGGACGAAGUUAUUAUGAAACAAUGAAUAAUUGAUAAUGUUAUUCAAUGCGAU-------------- ..(((((((((.......(((..(((((.((.........((((.(((((((....))))))).))))........)))))))..)))..))))))))).......-------------- ( -18.33, z-score = 0.22, R) >consensus ______AGCCUGCGCUUGCACUUAAGUUUUCCUUACAUUGUUGCGCCUUUUAUGGUGUGGCAUUAUUAUGAAACAGUGAAUAAUUGAUAAAGUUAUUCAAUGCAGUAAGUGACAUGCGGU .......((....((((((......(((((........(((..((((......))))..))).......)))))..(((((((((.....))))))))).....)))))).....))... (-20.64 = -21.84 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:09 2011