
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,686,552 – 7,686,663 |
| Length | 111 |
| Max. P | 0.654870 |

| Location | 7,686,552 – 7,686,663 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Shannon entropy | 0.48536 |
| G+C content | 0.48045 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -14.86 |
| Energy contribution | -14.55 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
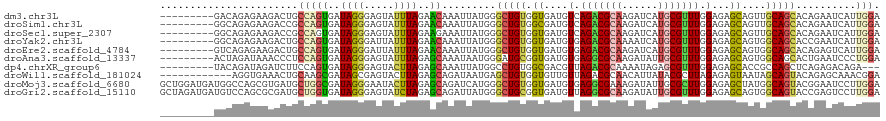
>dm3.chr3L 7686552 111 - 24543557 ---------GACAGAGAAGACUGCCAGUGAUAGGGAGUAUUUAGAACAAAUUAUGGGCUGUGGUGAUGUCAGACGCAAGAUCAUGCGUUUGGAGAGCAGUUGCAGCACAGAAUCAUUGGA ---------..(((......)))((((((((.....((.......))......((.((((..(((...(((((((((......)))))))))....)).)..)))).))..)))))))). ( -32.50, z-score = -2.43, R) >droSim1.chr3L 7185925 111 - 22553184 ---------GGCAGAGAAGACCGCCAGUGAUAGGGAGUAUUUAGAACAAAUUAUGGGCUGUGGCGAUGUCAGACGCAAGAUCAUGCGUUUGGAGAGCAGUUGCAGCACAGAAUCAUUGGA ---------(((......).)).((((((((.....((.......))......((.((((..((....(((((((((......)))))))))......))..)))).))..)))))))). ( -34.40, z-score = -2.73, R) >droSec1.super_2307 3161 111 - 3435 ---------GGCAGAGAAGACCGCCAGUGAUAGGGAGUAUUUAGAAGAAAUUAUGGGCUGUGGUGAUGUCAGACGCAAGAUCAUGCGUUUGGAGAGCAGUUGCAGCACAGAAUCAUUGGA ---------(((......).)).((((((((.............((....)).((.((((..(((...(((((((((......)))))))))....)).)..)))).))..)))))))). ( -32.70, z-score = -2.54, R) >droYak2.chr3L 8283469 111 - 24197627 ---------GGCAGAGAAGACUGCCAGUGAUAGGGAUUAUUUAGAACAAAUUAUGGGCUGUGGUGAUGUGAGACGCAAAAUCAUGCGUUUGGAGAGCAGUGGCAGCACCGAAUCAUUGGA ---------(((((......)))))((((((.((............((.....)).(((((.(((...(.(((((((......))))))).)....)).).))))).))..))))))... ( -31.70, z-score = -1.93, R) >droEre2.scaffold_4784 22100193 111 + 25762168 ---------GUCAGAGAAGACUGCCAGUGAUAGGGAUUAUUUAGAACAAAUUAUGGGCUGUGGUGAUGUGAGACGCAAGAUCAUGCGUUUGGAGAGCAGUGGCAGCACAGAGUCAUUGGA ---------(((......)))..((((((((...............((.....))..(((((.((.(((.(((((((......))))))).....)))....)).))))).)))))))). ( -29.30, z-score = -1.09, R) >droAna3.scaffold_13337 16478911 111 - 23293914 ---------ACUAGAUAAACCCUCCAGUGAUAGGGAGUAUUUAGAGCAAAUAAUGGGAUGCGGUGAUGUGAGGCGCAAGAUAUUGCGUUUGGAGAGCAGUGGCAGCACUGAAUCCCUGGA ---------.(((((((..((((........))))..)))))))..........(((((.(((((...(.((((((((....)))))))).)...((....))..))))).))))).... ( -36.10, z-score = -2.53, R) >dp4.chrXR_group6 1691841 108 - 13314419 ---------UACAGAUAGAUCUUCCAGUGAUAGGGAGUACUUAGAGCAAAUUAUGGCCUGUGGCGACGUUAGACGCAAAAUAGAGCGUUUGGAGAGCACCGCCAGCUCAGAGACAGA--- ---------...........(((((.......)))))..(((.((((......((((..(((.(....((((((((........))))))))...)))).)))))))).))).....--- ( -28.00, z-score = -1.09, R) >droWil1.scaffold_181024 791158 108 + 1751709 ------------AGGUGAAACUGCAAGCGAUAGCGAGUACUUAGAGCAGAUAAUGAGCUGUGGUGUUGUUAGACGCAACAUUAUACGCUUAGAGAGUAAUAGCAGUACAGAGCAAACGGA ------------.........(((..((....))..(((((............((((((((((((((((.....))))))))))).)))))............)))))...)))...... ( -26.75, z-score = -1.02, R) >droMoj3.scaffold_6680 17626210 120 + 24764193 GCUGGAUGAUGGCCAGCGUGAUGCUGGCGAUAGGGAAUACUUAGAGCAGAUCAUGGGCUGUGGUGAUGUGAGGCGAAAGAUAUUGCGCUUGGAGAGCUAUGGCAGUACGGAAUCCUUGGA ...((((..((..(..((((((.(((.(..((((.....))))..)))))))))).((((((((..(.((((((((......)))).)))).)..)))))))).)..))..))))..... ( -37.70, z-score = -1.34, R) >droGri2.scaffold_15110 7473587 120 + 24565398 GCUAGAUGAUGUCCAGCGCGAUGCUGGUGAUAGGGAGUAUCUAGAGCAGAUUAUGGGCUGCGGUGAUGUUAGGCGCAAGAUAUUGCGUUUGGAGAGCAGUGGCAGUACCGAGUCCUUGGA .(((((((.((((((((.....)))))..))).....)))))))..........(((((.(((((...((((((((((....))))))))))...((....))..))))))))))..... ( -38.60, z-score = -0.62, R) >consensus _________GGCAGAGAAGACUGCCAGUGAUAGGGAGUAUUUAGAACAAAUUAUGGGCUGUGGUGAUGUGAGACGCAAGAUCAUGCGUUUGGAGAGCAGUGGCAGCACAGAAUCAUUGGA .......................(((((..((((.....))))..)).........(((((.((....(.(((((((......))))))).)...))....)))))..........))). (-14.86 = -14.55 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:10:03 2011