| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,624,025 – 7,624,118 |
| Length | 93 |
| Max. P | 0.968463 |

| Location | 7,624,025 – 7,624,118 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.09 |
| Shannon entropy | 0.46237 |
| G+C content | 0.46512 |
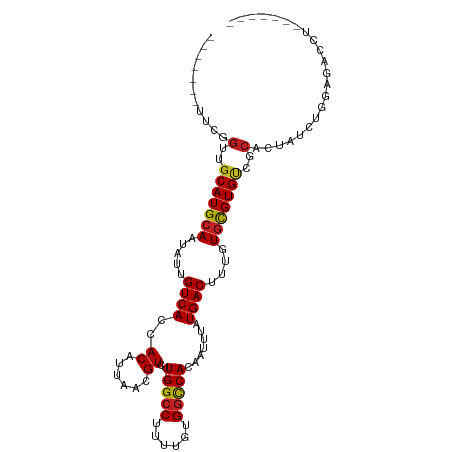
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -19.23 |
| Energy contribution | -19.22 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
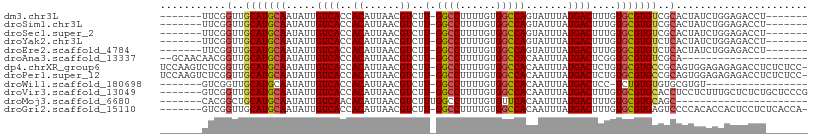
>dm3.chr3L 7624025 93 + 24543557 -------UUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCAGUAUUUAUGACUUUGUGCGUGUCGCACUAUCUGGAGACCU------- -------...(((.(((........))).)))........(((((-((((......))))...............(((((...)))))......)))))..------- ( -24.90, z-score = -1.02, R) >droSim1.chr3L 7138417 93 + 22553184 -------UUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCAGUAUUUAUGACUUUGUGCGUGUCGCACUAUCUGGAGACCU------- -------...(((.(((........))).)))........(((((-((((......))))...............(((((...)))))......)))))..------- ( -24.90, z-score = -1.02, R) >droSec1.super_2 7553173 93 + 7591821 -------UUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCAGUAUUUAUGACUUUGUGCGUGUCGCACUAUCUGGAGACCU------- -------...(((.(((........))).)))........(((((-((((......))))...............(((((...)))))......)))))..------- ( -24.90, z-score = -1.02, R) >droYak2.chr3L 8230215 93 + 24197627 -------UUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCAGUAUUUAUGACUUUGUGCGUGUCUCACUAUCUGGAGACCU------- -------...(((.(((((((.((..((((..((......)).((-((((......))))))......))))..)))))))))(((.(.....))))))).------- ( -25.30, z-score = -1.61, R) >droEre2.scaffold_4784 22048675 93 - 25762168 -------UUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCAGUAUUUAUGACUUUGUGCGUGUCUCACUAUCUGGAGACCU------- -------...(((.(((((((.((..((((..((......)).((-((((......))))))......))))..)))))))))(((.(.....))))))).------- ( -25.30, z-score = -1.61, R) >droAna3.scaffold_13337 16430198 84 + 23293914 --GCAACAACGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCACAAUUUAUGACUCGGUGCGUGUCGCA--------------------- --(((((....))))).(((.((((.(.((((............(-((((......)))))((.....))....))))))))).)))--------------------- ( -23.60, z-score = -0.65, R) >dp4.chrXR_group6 1635264 106 + 13314419 UCCAAGUCUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCACAAUUUAUGACUCUGUGCGUGCCGCAGUGGAGAGAGACCUCUCUCC- ...........((.(((((((.....((((..((......))..(-((((......))))).......))))....))))))).))...(((((((....)))))))- ( -34.90, z-score = -2.51, R) >droPer1.super_12 2325839 106 + 2414086 UCCAAGUCUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCACAAUUUAUGACUCUGUGCGUGCCGCAGUGGAGAGAGACCUCUCUCC- ...........((.(((((((.....((((..((......))..(-((((......))))).......))))....))))))).))...(((((((....)))))))- ( -34.90, z-score = -2.51, R) >droWil1.scaffold_180698 6195755 82 + 11422946 -------GUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCACAAUUUAUGACUCC-UCUGUGUGUGCGUGU----------------- -------.......(((((((.....((((..((......))..(-((((......))))).......))))...-....)))))))....----------------- ( -19.80, z-score = -0.26, R) >droVir3.scaffold_13049 5637228 100 + 25233164 -------GUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCACAAUUUAUGACUUUGUGCGUGCACCUCCUCUUUGCUCUCUGCUCCCG -------...(((.(((((((.((..((((..((......))..(-((((......))))).......))))..))))))))))))........((.....))..... ( -28.10, z-score = -3.44, R) >droMoj3.scaffold_6680 3980184 79 + 24764193 -------CACGGCUGCAUGCAAUAUUGUCAACACAUUAACGUCUUUGGCCUUUUGUGUUCACAAUUUAUGACUUUGUGCGUGCAGC---------------------- -------....((((((((((..(((((.((((((..((.((.....)).)).)))))).)))))...........))))))))))---------------------- ( -25.82, z-score = -2.78, R) >droGri2.scaffold_15110 11156247 99 - 24565398 -------GUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU-GGCCUUUUGUGGCCACAAUUUAUGACUUUGUGCGUGAGUCCCCACACCACUCCUCUCACCA- -------((.((...((((((.((..((((..((......))..(-((((......))))).......))))..))))))))....)).))................- ( -22.90, z-score = -0.98, R) >consensus _______UUCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUU_GGCCUUUUGUGGCCACAAUUUAUGACUUUGUGCGUGUCGCACUAUCUGGAGACCU_______ ...........(..(((((((.....((((..((......))....((((......))))........))))....)))))))..)...................... (-19.23 = -19.22 + -0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:59 2011