
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,592,283 – 7,592,333 |
| Length | 50 |
| Max. P | 0.977085 |

| Location | 7,592,283 – 7,592,333 |
|---|---|
| Length | 50 |
| Sequences | 10 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 86.18 |
| Shannon entropy | 0.26446 |
| G+C content | 0.44646 |
| Mean single sequence MFE | -12.83 |
| Consensus MFE | -10.62 |
| Energy contribution | -10.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7592283 50 + 24543557 GAAAAUCU-----GUUUAAUAUUUUCCCGCGC--ACUCACGCACGACGUGGGAAUUU--- ........-----..........((((((((.--..((......))))))))))...--- ( -11.30, z-score = -1.64, R) >droSim1.chr3L 7106704 50 + 22553184 GAAAAUCU-----GUUUAAUAUUUUCCCGCGC--ACUCACGCACGACGUGGGAAUUU--- ........-----..........((((((((.--..((......))))))))))...--- ( -11.30, z-score = -1.64, R) >droSec1.super_2 7521929 50 + 7591821 GAAAAUCU-----GUUUAAUAUUUUCCCGCGC--ACUCACGCACGACGUGGGAAUUU--- ........-----..........((((((((.--..((......))))))))))...--- ( -11.30, z-score = -1.64, R) >droYak2.chr3L 8198052 50 + 24197627 GAAAAUCU-----GUUUAAUAUUUUCCCGCGC--ACUCACGCACGACGUGGGAAUUU--- ........-----..........((((((((.--..((......))))))))))...--- ( -11.30, z-score = -1.64, R) >droEre2.scaffold_4784 22016992 50 - 25762168 GAAAAUCU-----GUUUAAUAUUUUCCCGCGC--ACUCACGCACGACGUGGGAAUUU--- ........-----..........((((((((.--..((......))))))))))...--- ( -11.30, z-score = -1.64, R) >dp4.chrXR_group6 11716668 52 - 13314419 GAAAAUCU-----AUUUAAUAUUUUCCCGCGCGCACUCACGCACGACGUGGGAAUUU--- ........-----..........((((((((((..(....)..)).))))))))...--- ( -13.30, z-score = -2.44, R) >droPer1.super_12 125833 52 - 2414086 GAAAAUCU-----AUUUAAUAUUUUCCCGCGCGCACUCACGCACGACGUGGGAAUUU--- ........-----..........((((((((((..(....)..)).))))))))...--- ( -13.30, z-score = -2.44, R) >droWil1.scaffold_180698 6154362 50 + 11422946 GAAAAUCU-----GUUUAAUAUUUUCCCGCGC--GCACACGCACGACGUGGGAAUUU--- ........-----..........(((((((((--(........)).))))))))...--- ( -14.00, z-score = -2.07, R) >droVir3.scaffold_13049 5601848 58 + 25233164 GAAAACUGUUUAAGUUUAAUAUUUUCCCGCGCG-C-GCACGCACAACGUGGGAAAAACUC ..(((((.....)))))....((((((((((.(-(-....))....)))))))))).... ( -17.50, z-score = -2.80, R) >droGri2.scaffold_15110 11122313 53 - 24565398 GAAAACUGUUUAAGUUUAAUAUUUUCCCGCGCG-CCGCACGCACAACGUGGGAA------ ..(((((.....)))))......((((((((.(-(.....))....))))))))------ ( -13.70, z-score = -1.13, R) >consensus GAAAAUCU_____GUUUAAUAUUUUCCCGCGC__ACUCACGCACGACGUGGGAAUUU___ .......................((((((((...............))))))))...... (-10.62 = -10.62 + 0.00)

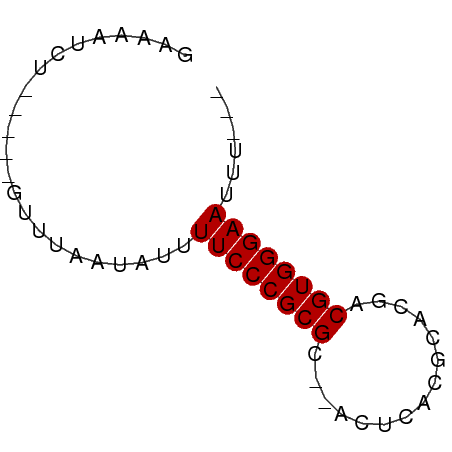
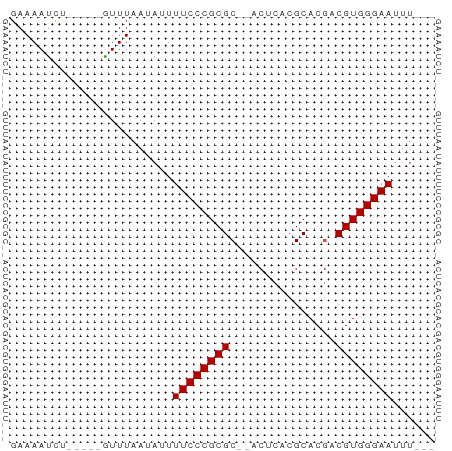
| Location | 7,592,283 – 7,592,333 |
|---|---|
| Length | 50 |
| Sequences | 10 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 86.18 |
| Shannon entropy | 0.26446 |
| G+C content | 0.44646 |
| Mean single sequence MFE | -15.29 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.30 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7592283 50 - 24543557 ---AAAUUCCCACGUCGUGCGUGAGU--GCGCGGGAAAAUAUUAAAC-----AGAUUUUC ---...(((((..(.((..(....).--.))))))))..........-----........ ( -14.20, z-score = -2.19, R) >droSim1.chr3L 7106704 50 - 22553184 ---AAAUUCCCACGUCGUGCGUGAGU--GCGCGGGAAAAUAUUAAAC-----AGAUUUUC ---...(((((..(.((..(....).--.))))))))..........-----........ ( -14.20, z-score = -2.19, R) >droSec1.super_2 7521929 50 - 7591821 ---AAAUUCCCACGUCGUGCGUGAGU--GCGCGGGAAAAUAUUAAAC-----AGAUUUUC ---...(((((..(.((..(....).--.))))))))..........-----........ ( -14.20, z-score = -2.19, R) >droYak2.chr3L 8198052 50 - 24197627 ---AAAUUCCCACGUCGUGCGUGAGU--GCGCGGGAAAAUAUUAAAC-----AGAUUUUC ---...(((((..(.((..(....).--.))))))))..........-----........ ( -14.20, z-score = -2.19, R) >droEre2.scaffold_4784 22016992 50 + 25762168 ---AAAUUCCCACGUCGUGCGUGAGU--GCGCGGGAAAAUAUUAAAC-----AGAUUUUC ---...(((((..(.((..(....).--.))))))))..........-----........ ( -14.20, z-score = -2.19, R) >dp4.chrXR_group6 11716668 52 + 13314419 ---AAAUUCCCACGUCGUGCGUGAGUGCGCGCGGGAAAAUAUUAAAU-----AGAUUUUC ---...(((((.((.((..(....)..)))).)))))..........-----........ ( -19.80, z-score = -3.96, R) >droPer1.super_12 125833 52 + 2414086 ---AAAUUCCCACGUCGUGCGUGAGUGCGCGCGGGAAAAUAUUAAAU-----AGAUUUUC ---...(((((.((.((..(....)..)))).)))))..........-----........ ( -19.80, z-score = -3.96, R) >droWil1.scaffold_180698 6154362 50 - 11422946 ---AAAUUCCCACGUCGUGCGUGUGC--GCGCGGGAAAAUAUUAAAC-----AGAUUUUC ---...(((((.((.((((....)))--))).)))))..........-----........ ( -13.40, z-score = -1.32, R) >droVir3.scaffold_13049 5601848 58 - 25233164 GAGUUUUUCCCACGUUGUGCGUGC-G-CGCGCGGGAAAAUAUUAAACUUAAACAGUUUUC ....(((((((.(((.(((....)-)-)))).)))))))....(((((.....))))).. ( -16.00, z-score = -1.50, R) >droGri2.scaffold_15110 11122313 53 + 24565398 ------UUCCCACGUUGUGCGUGCGG-CGCGCGGGAAAAUAUUAAACUUAAACAGUUUUC ------(((((.....(((((.....-))))))))))......(((((.....))))).. ( -12.90, z-score = -0.48, R) >consensus ___AAAUUCCCACGUCGUGCGUGAGU__GCGCGGGAAAAUAUUAAAC_____AGAUUUUC ......(((((.(((...((....))..))).)))))....................... (-13.07 = -13.30 + 0.23)

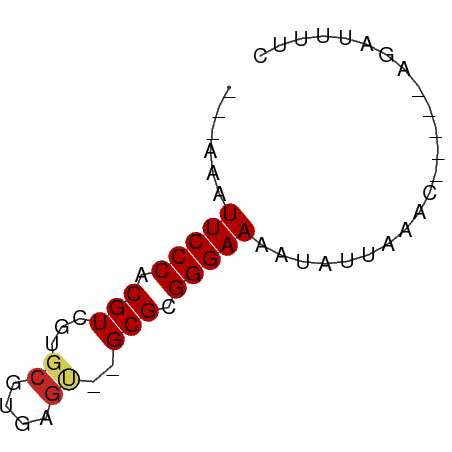
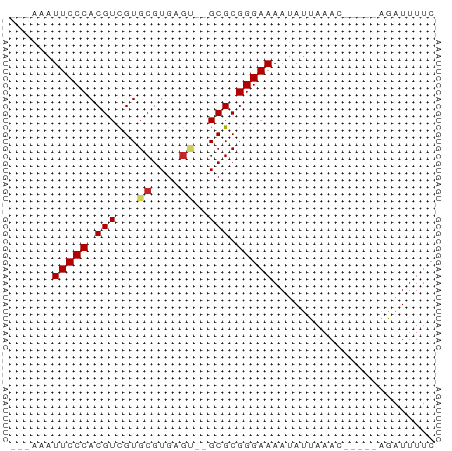
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:53 2011