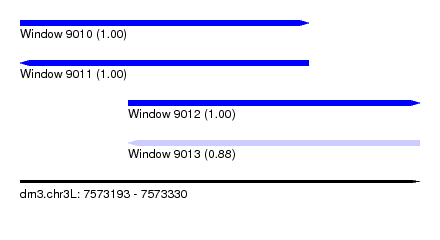
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,573,193 – 7,573,330 |
| Length | 137 |
| Max. P | 0.998449 |

| Location | 7,573,193 – 7,573,292 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.49 |
| Shannon entropy | 0.18551 |
| G+C content | 0.29156 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -20.22 |
| Energy contribution | -19.23 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.89 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.998449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7573193 99 + 24543557 AAAAGGCCAGAACUGACAAGAACAAUAACUUGUUUACAAAACUAAUCCUAUUAACUUAGGAUUAGGAAAGCAUAAGAAUCUAUUAUUUUUCUUCUUGUU ..............((((((((...................(((((((((......)))))))))(((((.((((.......))))))))))))))))) ( -21.10, z-score = -2.89, R) >droSim1.chr3L 7087425 98 + 22553184 AAAAGACCAGAACUAACAAGAACAAUAAACUCUUUACAAA-CCAAUCCUAUAAUAUUAGGAUUGGGGUAUCAUAAGAAUCUAUUAGUUCUUUUCUUGUU ..((((..((((((((..(((.............(((...-(((((((((......))))))))).)))((....)).))).))))))))..))))... ( -26.60, z-score = -4.83, R) >droSec1.super_2 7502780 98 + 7591821 AAGAGACCAGAACUGACAAAAACAAUAAACUCUUUACAAA-CCAAUCCUAUAAUCUUAGGAUUGGGAUAUCAUAAGAACCUAUUAGUUCUUUUCUUGUU ..((((..((((((((........................-(((((((((......)))))))))....((....)).....))))))))..))))... ( -24.40, z-score = -3.94, R) >consensus AAAAGACCAGAACUGACAAGAACAAUAAACUCUUUACAAA_CCAAUCCUAUAAUCUUAGGAUUGGGAUAUCAUAAGAAUCUAUUAGUUCUUUUCUUGUU ..((((..((((((((.........................(((((((((......)))))))))(....)...........))))))))..))))... (-20.22 = -19.23 + -0.99)

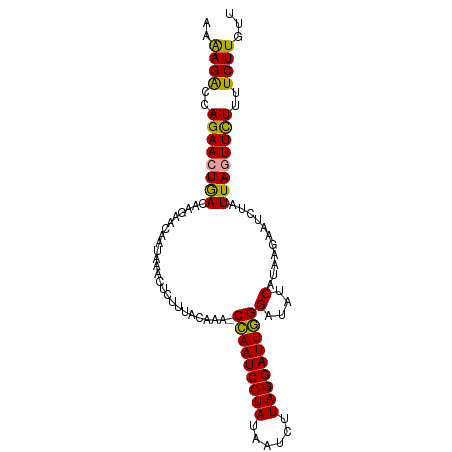
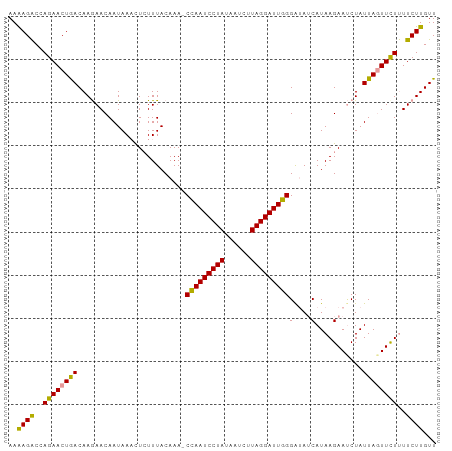
| Location | 7,573,193 – 7,573,292 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.49 |
| Shannon entropy | 0.18551 |
| G+C content | 0.29156 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -21.49 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.997790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7573193 99 - 24543557 AACAAGAAGAAAAAUAAUAGAUUCUUAUGCUUUCCUAAUCCUAAGUUAAUAGGAUUAGUUUUGUAAACAAGUUAUUGUUCUUGUCAGUUCUGGCCUUUU ......(((((.((((((................(((((((((......)))))))))..(((....))))))))).)))))((((....))))..... ( -19.30, z-score = -1.66, R) >droSim1.chr3L 7087425 98 - 22553184 AACAAGAAAAGAACUAAUAGAUUCUUAUGAUACCCCAAUCCUAAUAUUAUAGGAUUGG-UUUGUAAAGAGUUUAUUGUUCUUGUUAGUUCUGGUCUUUU ...((((..(((((((((((((((((...(((..(((((((((......)))))))))-..))).))))))).........))))))))))..)))).. ( -32.20, z-score = -5.61, R) >droSec1.super_2 7502780 98 - 7591821 AACAAGAAAAGAACUAAUAGGUUCUUAUGAUAUCCCAAUCCUAAGAUUAUAGGAUUGG-UUUGUAAAGAGUUUAUUGUUUUUGUCAGUUCUGGUCUCUU ....(((..((((((((((..(((((...(((..(((((((((......)))))))))-..))).)))))..)))).........))))))..)))... ( -29.20, z-score = -4.07, R) >consensus AACAAGAAAAGAACUAAUAGAUUCUUAUGAUAUCCCAAUCCUAAGAUUAUAGGAUUGG_UUUGUAAAGAGUUUAUUGUUCUUGUCAGUUCUGGUCUUUU .(((((((......((((((((((((((((....(((((((((......)))))))))..)))).)))))))))))))))))))............... (-21.49 = -21.50 + 0.01)


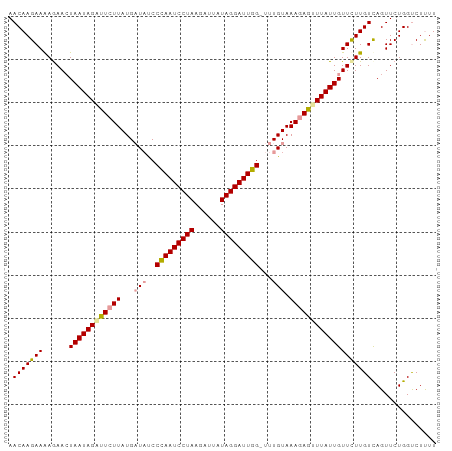
| Location | 7,573,230 – 7,573,330 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 72.56 |
| Shannon entropy | 0.34895 |
| G+C content | 0.27615 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -16.13 |
| Energy contribution | -15.69 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998078 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 7573230 100 + 24543557 AAAACUAAUCCUAUUAACUUAGGAUUAGGAAAGCAUAAGAAUCUAUUAUUUUUCUUCUUGUUUAGCCAGUCUUUCAAAGAAUUUUAAUCUAGACCUUUGU ....(((((((((......)))))))))..(((((.(((((..........)))))..))))).....((((.....(((.......)))))))...... ( -18.30, z-score = -1.23, R) >droSim1.chr3L 7087462 77 + 22553184 -AAACCAAUCCUAUAAUAUUAGGAUUGGGGUAUCAUAAGAAUCUAUUAGUUCUUUUCUUGUUUAGCCAUUCUUUUAAA---------------------- -...(((((((((......)))))))))(((...(((((((.............)))))))...)))...........---------------------- ( -21.12, z-score = -4.19, R) >droSec1.super_2 7502817 77 + 7591821 -AAACCAAUCCUAUAAUCUUAGGAUUGGGAUAUCAUAAGAACCUAUUAGUUCUUUUCUUGUUUAGCCAUACUUUUAAA---------------------- -...(((((((((......)))))))))((((....((((((......))))))....))))................---------------------- ( -19.50, z-score = -3.80, R) >consensus _AAACCAAUCCUAUAAUCUUAGGAUUGGGAUAUCAUAAGAAUCUAUUAGUUCUUUUCUUGUUUAGCCAUUCUUUUAAA______________________ ....(((((((((......)))))))))......(((((((.............)))))))....................................... (-16.13 = -15.69 + -0.44)

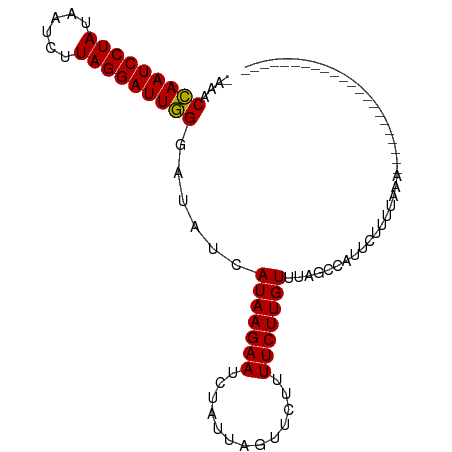
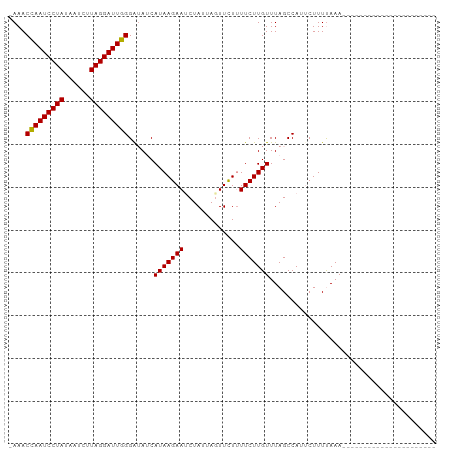
| Location | 7,573,230 – 7,573,330 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 72.56 |
| Shannon entropy | 0.34895 |
| G+C content | 0.27615 |
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -12.66 |
| Energy contribution | -12.22 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879669 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 7573230 100 - 24543557 ACAAAGGUCUAGAUUAAAAUUCUUUGAAAGACUGGCUAAACAAGAAGAAAAAUAAUAGAUUCUUAUGCUUUCCUAAUCCUAAGUUAAUAGGAUUAGUUUU .((((((.............))))))...((..(((.....(((((.............)))))..))).))(((((((((......))))))))).... ( -16.74, z-score = -0.30, R) >droSim1.chr3L 7087462 77 - 22553184 ----------------------UUUAAAAGAAUGGCUAAACAAGAAAAGAACUAAUAGAUUCUUAUGAUACCCCAAUCCUAAUAUUAUAGGAUUGGUUU- ----------------------.(((.((((((..(((.................))))))))).)))....(((((((((......)))))))))...- ( -13.93, z-score = -1.79, R) >droSec1.super_2 7502817 77 - 7591821 ----------------------UUUAAAAGUAUGGCUAAACAAGAAAAGAACUAAUAGGUUCUUAUGAUAUCCCAAUCCUAAGAUUAUAGGAUUGGUUU- ----------------------........................(((((((....)))))))........(((((((((......)))))))))...- ( -18.30, z-score = -2.56, R) >consensus ______________________UUUAAAAGAAUGGCUAAACAAGAAAAGAACUAAUAGAUUCUUAUGAUAUCCCAAUCCUAAGAUUAUAGGAUUGGUUU_ .........................................(((((.............)))))........(((((((((......))))))))).... (-12.66 = -12.22 + -0.44)

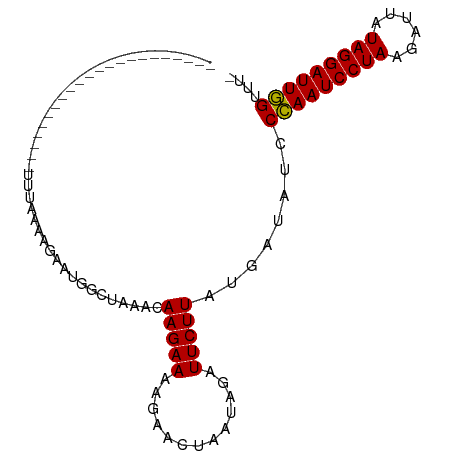
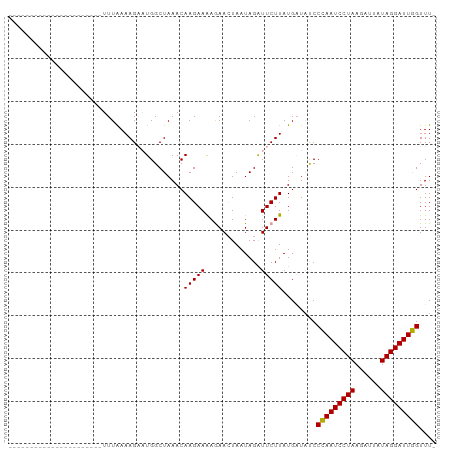
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:51 2011