
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,565,765 – 7,565,839 |
| Length | 74 |
| Max. P | 0.944136 |

| Location | 7,565,765 – 7,565,839 |
|---|---|
| Length | 74 |
| Sequences | 11 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Shannon entropy | 0.50301 |
| G+C content | 0.47321 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -13.53 |
| Energy contribution | -13.71 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903783 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
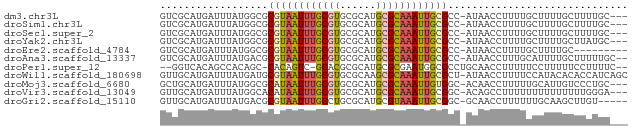
>dm3.chr3L 7565765 74 + 24543557 GUCGCAUGAUUUAUGGCGCGUAAUUUGCGUGCGCAUGCGCAAAUUGCGCC-AUAACCUUUUGCUUUUGCUUUUGC--- ...(((....((((((((((..((((((((......))))))))))))))-)))).....)))............--- ( -27.50, z-score = -2.38, R) >droSim1.chr3L 7080135 74 + 22553184 GUCGCAUGAUUUAUGGCGCGUAAUUUGCGUGCGCAUGCGCAAAUUGCGCC-AUAACCUUUUGCUUUUGCUUUUGC--- ...(((....((((((((((..((((((((......))))))))))))))-)))).....)))............--- ( -27.50, z-score = -2.38, R) >droSec1.super_2 7495552 74 + 7591821 GUCGCAUGAUUUAUGGCGCGUAAUUUGCGUGCGCAUGCGCAAAUUGCGCC-AUAACCUUUUGCUUUUGCUUUUGC--- ...(((....((((((((((..((((((((......))))))))))))))-)))).....)))............--- ( -27.50, z-score = -2.38, R) >droYak2.chr3L 8170180 74 + 24197627 GUCGCAUGAUUUAUGGCGCGUAAUUUGCGUGCGCAUGCGCAAAUUGCGCC-AUAACCUUUUGCUUUUGCUUAUGC--- ...((((((.((((((((((..((((((((......))))))))))))))-))))......((....))))))))--- ( -30.10, z-score = -3.25, R) >droEre2.scaffold_4784 21990030 68 - 25762168 GUCGCAUGAUUUAUGGCGCGUAAUUUGCGUGCGCAUGCGCAAAUUGCGCC-AUAACCUUUUGCUUUUGC--------- ...(((....((((((((((..((((((((......))))))))))))))-)))).....)))......--------- ( -27.50, z-score = -2.88, R) >droAna3.scaffold_13337 16370034 75 + 23293914 GUCGCAUGAUUUAUGACGCGUAAUUUGCGUGCGCAUGCGCAAAUUGCGCC-AUAACCUUUGCAUUUUGCUUUUUGC-- ...(((....(((((..(((((((((((((......))))))))))))))-))))....)))..............-- ( -25.90, z-score = -2.27, R) >droPer1.super_12 2258383 72 + 2414086 --GGUCACAGCCACAGC-CACAGUC-GCACGCGCAUGCGCGAAUGGCGCCUGCAACCUUUUUCCUUUUUCCUUUUC-- --(((..(((.....((-((...((-((.((....)).)))).))))..)))..)))...................-- ( -17.20, z-score = -0.68, R) >droWil1.scaffold_180698 5324700 77 - 11422946 GUUGCAUGAUUUAUGAUGCGUAAUUUGCGUGCGCAAGCGCAAAUUGCGCU-AUAACCUUUUCCAUACACACCAUCAGC ((((.(((...((((..(((((((((((((......))))))))))))).-...........)))).....))))))) ( -21.24, z-score = -1.57, R) >droMoj3.scaffold_6680 3911204 74 + 24764193 GCUGCAUGAUUUAUGGCGCAUAAUUUGCGUGCGCAUGCGCAAAUUGUGGC-ACAACCUUUUUGCAUUGUCCCUGC--- ..((((.((....((((.((((((((((((......))))))))))))))-.))....)).))))..........--- ( -22.90, z-score = -0.34, R) >droVir3.scaffold_13049 5569231 74 + 25233164 GUUGCAUGAUUUAUGGCACAUAAUUUGCGUGCGCAUGCGCAAAUUGCGGC-ACAGCCUUUUUUUUUUUUUUGGGA--- ((((((....(((((...)))))(((((((......))))))).))))))-....(((.............))).--- ( -15.92, z-score = 1.04, R) >droGri2.scaffold_15110 11095522 72 - 24565398 GUUGCAUGAUUUAUGACGCGUAAUUUGCCUGCGCAUGCGUAAAUUGCGGC-GCAACCUUUUUUGCAAGCUUGU----- ((((((..(((((((..(((((.......)))))...)))))))))))))-((((......))))........----- ( -20.70, z-score = -0.28, R) >consensus GUCGCAUGAUUUAUGGCGCGUAAUUUGCGUGCGCAUGCGCAAAUUGCGCC_AUAACCUUUUGCUUUUGCUUUUGC___ ..................((((((((((((......)))))))))))).............................. (-13.53 = -13.71 + 0.18)

| Location | 7,565,765 – 7,565,839 |
|---|---|
| Length | 74 |
| Sequences | 11 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Shannon entropy | 0.50301 |
| G+C content | 0.47321 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -12.47 |
| Energy contribution | -13.35 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944136 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
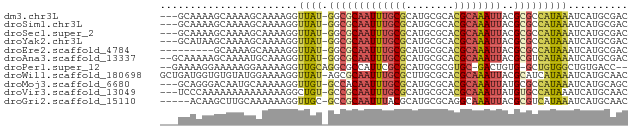
>dm3.chr3L 7565765 74 - 24543557 ---GCAAAAGCAAAAGCAAAAGGUUAU-GGCGCAAUUUGCGCAUGCGCACGCAAAUUACGCGCCAUAAAUCAUGCGAC ---(((...((....))....((((((-(((((((((((((........))))))))..))))))).)))).)))... ( -27.20, z-score = -3.29, R) >droSim1.chr3L 7080135 74 - 22553184 ---GCAAAAGCAAAAGCAAAAGGUUAU-GGCGCAAUUUGCGCAUGCGCACGCAAAUUACGCGCCAUAAAUCAUGCGAC ---(((...((....))....((((((-(((((((((((((........))))))))..))))))).)))).)))... ( -27.20, z-score = -3.29, R) >droSec1.super_2 7495552 74 - 7591821 ---GCAAAAGCAAAAGCAAAAGGUUAU-GGCGCAAUUUGCGCAUGCGCACGCAAAUUACGCGCCAUAAAUCAUGCGAC ---(((...((....))....((((((-(((((((((((((........))))))))..))))))).)))).)))... ( -27.20, z-score = -3.29, R) >droYak2.chr3L 8170180 74 - 24197627 ---GCAUAAGCAAAAGCAAAAGGUUAU-GGCGCAAUUUGCGCAUGCGCACGCAAAUUACGCGCCAUAAAUCAUGCGAC ---((((..((....))....((((((-(((((((((((((........))))))))..))))))).))))))))... ( -28.20, z-score = -3.62, R) >droEre2.scaffold_4784 21990030 68 + 25762168 ---------GCAAAAGCAAAAGGUUAU-GGCGCAAUUUGCGCAUGCGCACGCAAAUUACGCGCCAUAAAUCAUGCGAC ---------......(((.....((((-(((((((((((((........))))))))..)))))))))....)))... ( -26.00, z-score = -3.29, R) >droAna3.scaffold_13337 16370034 75 - 23293914 --GCAAAAAGCAAAAUGCAAAGGUUAU-GGCGCAAUUUGCGCAUGCGCACGCAAAUUACGCGUCAUAAAUCAUGCGAC --((.....))....((((....((((-(((((((((((((........))))))))..)))))))))....)))).. ( -25.00, z-score = -2.40, R) >droPer1.super_12 2258383 72 - 2414086 --GAAAAGGAAAAAGGAAAAAGGUUGCAGGCGCCAUUCGCGCAUGCGCGUGC-GACUGUG-GCUGUGGCUGUGACC-- --...................(((..(((.((((((((((((......))))-))..)))-))...).)))..)))-- ( -27.30, z-score = -1.97, R) >droWil1.scaffold_180698 5324700 77 + 11422946 GCUGAUGGUGUGUAUGGAAAAGGUUAU-AGCGCAAUUUGCGCUUGCGCACGCAAAUUACGCAUCAUAAAUCAUGCAAC ((((((((((((((.............-.((((.....))))((((....))))..)))))))))....))).))... ( -23.50, z-score = -1.25, R) >droMoj3.scaffold_6680 3911204 74 - 24764193 ---GCAGGGACAAUGCAAAAAGGUUGU-GCCACAAUUUGCGCAUGCGCACGCAAAUUAUGCGCCAUAAAUCAUGCAGC ---(((((.(((((........)))))-.)).......((((((((....))).....))))).........)))... ( -19.70, z-score = 0.53, R) >droVir3.scaffold_13049 5569231 74 - 25233164 ---UCCCAAAAAAAAAAAAAAGGCUGU-GCCGCAAUUUGCGCAUGCGCACGCAAAUUAUGUGCCAUAAAUCAUGCAAC ---..................((((((-(((((.....)))...))))).(((.....)))))).............. ( -13.30, z-score = 0.59, R) >droGri2.scaffold_15110 11095522 72 + 24565398 -----ACAAGCUUGCAAAAAAGGUUGC-GCCGCAAUUUACGCAUGCGCAGGCAAAUUACGCGUCAUAAAUCAUGCAAC -----....((((((......((....-.))(((.........))))))))).......((((........))))... ( -16.80, z-score = 0.23, R) >consensus ___GCAAAAGCAAAAGCAAAAGGUUAU_GGCGCAAUUUGCGCAUGCGCACGCAAAUUACGCGCCAUAAAUCAUGCGAC .....................((((...(((((((((((((........))))))))..)))))...))))....... (-12.47 = -13.35 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:48 2011