| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,557,480 – 7,557,591 |
| Length | 111 |
| Max. P | 0.657028 |

| Location | 7,557,480 – 7,557,591 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.29 |
| Shannon entropy | 0.80316 |
| G+C content | 0.51593 |
| Mean single sequence MFE | -35.43 |
| Consensus MFE | -11.63 |
| Energy contribution | -11.72 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
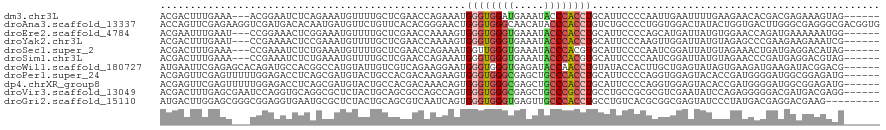
>dm3.chr3L 7557480 111 + 24543557 ACGACUUUGAAA---ACGGAAUCUCAGAAAUGUUUUGCUCGAACCAGAAAUGGGUGGAUGAAAUACCCACCUGCAUUCCCCAAUUGAAUUUUGAAGAACACGACGAGAAAGUAG------ ...(((((.(((---(((............)))))).((((..........((((((.........))))))...(((..(((.......)))..))).....)))))))))..------ ( -18.30, z-score = 0.52, R) >droAna3.scaffold_13337 10546789 120 + 23293914 ACCAGUUCGAGAAGGUCGAUGACACAAUGAUGUUCUGUUCACACGGGAACUGGGUGGGCAACAUACCCACCUGUCUGCCCCUGGUGGACUAUACUGGUGACUUGGGCGAGGGCGACGGUG (((.((((.((((.((((.((...)).)))).))))((((((((.((....(((((((.......)))))))(((..((...))..)))....)).)))...)))))..))))...))). ( -40.90, z-score = 0.10, R) >droEre2.scaffold_4784 21981410 111 - 25762168 ACGAAUUUGAAU---CCGGAAACUCGGAAAUGUUUUGCUCGAACCAAAAGUGGGUGGGUGAAAUACCCACCUGCAUUCCCCAGCAUGAUUAUGUGGAACCAGAUGAAAAAAUGG------ ....(((((..(---(((....)..((((...(((((.......)))))(..((((((((...))))))))..).))))...((((....)))))))..)))))..........------ ( -29.60, z-score = -1.22, R) >droYak2.chr3L 8161583 111 + 24197627 ACGACUUUGAAU---CCGAAAACUCCGAAAUGUUUUGCUCGAACCAAAAGUGGGUGGGUGAAAUACCCACCUGCAUUCCCAAGUUGGAUUAUGUAGAGCCCGAAGAAGAAAUCG------ .(..(((((...---...(((((........)))))((((.........(..((((((((...))))))))..)..(((......))).......)))).)))))..)......------ ( -31.10, z-score = -2.63, R) >droSec1.super_2 7487007 111 + 7591821 ACGACUUUGAAA---CCGAAAUCUCUGAAAUGUUUUGCUCGAACCAGAAAUGGUUGGGUGAAAUACCCACGUGCAUUCCCCAAUCGGAUUAUGUAGAAACUGAUGAGGACAUAG------ ..((.((((...---.)))).))......((((..(((.(((((((....)))))(((((...))))).)).)))...((..(((((.((......)).)))))..))))))..------ ( -26.20, z-score = -1.33, R) >droSim1.chr3L 7071573 111 + 22553184 ACGACUUUGAAA---CCGAAAUCUCUGAAAUGUUUUGCUCGAACCAGAAAUGGGUGGGUGAAAUACCCACGUGCAUUCCCCAAUCGGAUUAUGUAGAACCCGAUGAGGACGUAG------ ..((.((((...---.)))).))......((((..(((.(...(((....)))(((((((...)))))))).)))...((..(((((.((......)).)))))..))))))..------ ( -28.40, z-score = -1.07, R) >droWil1.scaffold_180727 2571840 114 + 2741493 AUGAAUUCGAGAGCACAGAUGCCACGGCCAUGUAUUGUCGUCAGAAGGAAUGGGUGGGUGAGAUACCAACCUGUAUACCACUUGCUGAGUAUAGUGAAGAUGAAGAUACGGACG------ ......(((.(.(((....)))).)))...((((((.(((((....((.((((((.((((...)))).))))))...))..(..(((....)))..).))))).))))))....------ ( -25.60, z-score = 0.47, R) >droPer1.super_24 1306477 114 + 1556852 ACGAGUUCGAGUUUUUGGAGACCUCAGCGAUGUACUGCCACGACAAGAAGUGGGUGGGCGAGCUGCCCACCUGCAUUCCCCAGGUGGAGUACACCGAUGGGGAUGGCGGAGAUG------ (((.(((.(((.....(....)))))))..))).((((((.........(..(((((((.....)))))))..)..((((((((((.....))))..)))))))))))).....------ ( -48.50, z-score = -3.01, R) >dp4.chrXR_group8 7468985 114 - 9212921 ACGAGUUCGAGUUUUUGGAGACCUCAGCGAUGUACUGCCACGACAAACAGUGGGUGGGCGAGCUGCCCACCUGCAUUCCCCAGGUGGAGUACACCGAUGGGGAUGGCGGAGAUG------ (((.(((.(((.....(....)))))))..))).((((((.........(..(((((((.....)))))))..)..((((((((((.....))))..)))))))))))).....------ ( -48.50, z-score = -2.91, R) >droVir3.scaffold_13049 20541747 114 + 25233164 ACGACUUUGAGCGAAUCCAGGUGCAGGCGCUCUACUGCAGCGCCAGCCAGUGGGUGGGCGAGCUGCCCGCCUGCCUGCCGCGCGUCGAAUAUCCAGAGGGGGACGAUGACGAGG------ ....(((((.(((....(((((((.(((((((....).)))))).))....((((((((.....))))))))))))).))).(((((....(((....)))..))))).)))))------ ( -47.90, z-score = -0.03, R) >droGri2.scaffold_15110 12597355 111 - 24565398 AUGACUUGGAGCGGGCGGAGGUGAAUGCGCUCUACUGCAGCGUCAAUCAGUGGGUGGGUGAGUUGCCCACCUGCCUGUCACGCGGCGAGUAUCCCUAUGACGAGGACGAAG--------- ...(((((..((((((((.(((..((((((......)).))))..))).(..(((((((.....)))))))..)))))).)))..)))))...(((......)))......--------- ( -44.70, z-score = -1.26, R) >consensus ACGACUUUGAGA___CCGAAAUCUCAGAAAUGUUUUGCUCGAACAAGAAGUGGGUGGGUGAAAUACCCACCUGCAUUCCCCAGGUGGAUUAUGCAGAAGACGAUGACGAAGUAG______ ...................................................((((((((.....))))))))................................................ (-11.63 = -11.72 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:46 2011