| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,567,670 – 4,567,766 |
| Length | 96 |
| Max. P | 0.838299 |

| Location | 4,567,670 – 4,567,766 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.89 |
| Shannon entropy | 0.52885 |
| G+C content | 0.59513 |
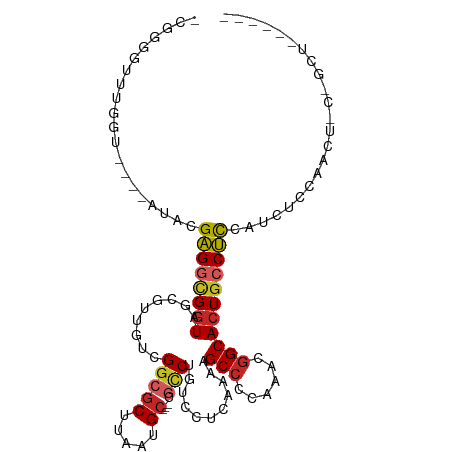
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.63 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
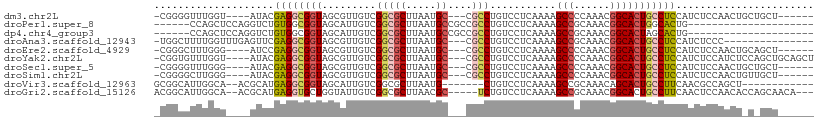
>dm3.chr2L 4567670 96 - 23011544 -CGGGGUUUGGU----AUACGAGGCGGUAGCGUUGUCGGCGCUUAAUGC---CGCCUGUCCUCAAAAGCCCCAAACGGCACUGCCUCCAUCUCCAACUGCUGCU------ -.(((((((...----..((.((((((((((((.....)))))....))---)))))))......)))))))...(((((.................)))))..------ ( -33.83, z-score = -0.79, R) >droPer1.super_8 1451835 83 + 3966273 ------CCAGCUCCAGGUCUGUGGCGGUAGCAUUGUCGGCGCUUAAUGCCGCCGCCUGUCCUCAAAAGCCGCAAACGGCACUGGCACUG--------------------- ------.(((..((((......(((((..((((((........))))))..)))))...........((((....)))).))))..)))--------------------- ( -34.20, z-score = -1.86, R) >dp4.chr4_group3 405182 83 + 11692001 ------CCAGCUCCAGGUCUGUGGCGGUAGCAUUGUCGGCGCUUAAUGCCGCCGCCUGUCCUCAAAAGCCGCAAACGGCACUAGCACUG--------------------- ------...(((..(((.(...(((((..((((((........))))))..))))).).))).....((((....))))...)))....--------------------- ( -32.10, z-score = -1.72, R) >droAna3.scaffold_12943 1766514 91 + 5039921 -UGGCUUUUGGUUUGAGUUCGAGGCGGUAGCGUUGUCGGCGCUUAAUGC---CGCCUGUCCUCAAAAGCCGCAAACGGCACUGCCUCCAUCUCCC--------------- -.(((......((((((..(.((((((((((((.....)))))....))---))))))..)))))).((((....))))...)))..........--------------- ( -36.70, z-score = -2.65, R) >droEre2.scaffold_4929 4651138 96 - 26641161 -CGGGCUUUGGG----AUCCGAGGCGGUAGCGUUGUCGGCGCUUAAUGC---CGCCUGUCCUCAAAAGCCCCAAACGGCACUGCCUCCAUCUCCAACUGCAGCU------ -.((((((((((----((...((((((((((((.....)))))....))---))))))))))..))))))).....(((...)))...................------ ( -36.50, z-score = -1.34, R) >droYak2.chr2L 4585296 102 - 22324452 -CGGUGUUUGGU----AUACGAGGCGGUAGCGUUGUCGGCGCUUAAUGC---CGCCUGUCCUCAAAAGCCCCAAACGGCACUGCCUCCAUCUCCAUCUCCAGCUGCAGCU -.((.(..(((.----....(((((((((((((.....)))))....((---((..((..((....))...))..)))))))))))).....))).).))(((....))) ( -32.20, z-score = -0.46, R) >droSec1.super_5 2660516 96 - 5866729 -CGGGGUUUGGG----AUACGAGGCGGUAGCGUUGUCGGCGCUUAAUGC---CGCCUGUCCUCAAAAGCCCCAAACGGCACUGCCUCCAUCUCCAACUGCUGCU------ -.((((((((((----((...((((((((((((.....)))))....))---))))))))))...)))))))...(((((.................)))))..------ ( -37.53, z-score = -1.52, R) >droSim1.chr2L 4453587 96 - 22036055 -CGGGGCUUGGG----AUACGAGGCGGUAGCGUUGUCGGCGCUUAAUGC---CGCCUGUCCUCAAAAGCCCCAAACGGCACUGCCUCCAUCUCCAACUGUUGCU------ -.((((((((((----((...((((((((((((.....)))))....))---))))))))))...)))))))....(((...)))...................------ ( -39.40, z-score = -2.10, R) >droVir3.scaffold_12963 8478570 89 + 20206255 GCGGCAUUGGCA--ACGCAUGAGGCGGUAGCAUUGUCGGCGCUUAAUG-------CUGUCCUCAAAAGCCGCAAACAGCACUGCCUUCAACGCCAGCU------------ (((((...(...--.)...(((((((((.((.(((.((((((.....)-------)((....))...)))))))...)))))))).)))..))).)).------------ ( -31.10, z-score = -0.33, R) >droGri2.scaffold_15126 3119357 100 + 8399593 ACGGCAUUGGCA--ACGCAUGAGGUGCUGGUAUUGUCGGCGCUUAACGC-----UCUGUCCUCAAAAGCCGCAAACGGCACUGCCUUCAACUCCAACACCAGCAACA--- ...((...(...--.)......((((.(((..(((..(((((.....))-----.............((((....))))...)))..)))..))).)))).))....--- ( -29.90, z-score = -0.52, R) >consensus _CGGGGUUUGGU____AUACGAGGCGGUAGCGUUGUCGGCGCUUAAUGC___CGCCUGUCCUCAAAAGCCCCAAACGGCACUGCCUCCAUCUCCAACU_C_GCU______ ....................((((((((.((((((........))))))..................(((......)))))))))))....................... (-18.76 = -18.63 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:44 2011