
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,538,168 – 7,538,276 |
| Length | 108 |
| Max. P | 0.561687 |

| Location | 7,538,168 – 7,538,276 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.05 |
| Shannon entropy | 0.58728 |
| G+C content | 0.44083 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -12.18 |
| Energy contribution | -13.32 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7538168 108 + 24543557 ------UGGGGCGGCACUGCAGUGCGACGUUGGAAU-UGGUGGAGCUGCAGUUGAAAAGCGACUGUCAAUGAAAAUGAAAUUUUAUUGAGAUUUUUA-CCGAAACCCCCUGCUUCU- ------.((((.(((((....))))..........(-((((((((...((((((.....))))))(((((((((......)))))))))...)))))-))))..))))).......- ( -32.70, z-score = -1.85, R) >droAna3.scaffold_13337 16348979 95 + 23293914 ---------------------GCGGACUG-CAGUCUGUUGGGGAGUUGCAGUUGAAAAGCGGCUGUCAAUGAAAAUGAAACUUUAUUGAGAUUUUUGUCCCCCUCCUCGAUGCUCCU ---------------------((((((..-..))))(((((((((...((((((.....))))))((((((((........)))))))).............))))))))))).... ( -27.40, z-score = -1.34, R) >droEre2.scaffold_4784 21961518 108 - 25762168 ------UGGGGCGGCAAUGCAACGCGAUGGUGGAAU-UGGUGGAGCUGCAGUUGAAAAGCGGCUGUCAAUGAAAAUGAAAUUUUAUUGAGAUUUUUA-CCGAAUCCCCCUGCUUCU- ------.(((((((...(((...)))..((.(((.(-((((((((((((.........)))))(.(((((((((......))))))))).)..))))-)))).)))))))))))).- ( -35.40, z-score = -2.08, R) >droYak2.chr3L 8142008 109 + 24197627 ------UGGGGCGGCACUGAACCGCGAUGUUGGAAU-UGGUGGAGCUGCAGUUGAAAAGCGGCUGUCAAUGAAAAUGAAAUUUUAUUGAGAUUUUUUUCCGAAACCCCCUGCUACU- ------.(((((((.......))))....((((((.-......((((((.........)))))).(((((((((......))))))))).......))))))....))).......- ( -30.40, z-score = -1.05, R) >droSec1.super_2 7467758 108 + 7591821 ------UGGUGCGGCACUGCAGCGCGAUGUUGGAAU-UGGUGGAGCUGCAGUUGAAAAGCGGCUGUCAAUGAAAAUGAAAUUUUAUUGAGAUUUUUA-CCGAAACCCCCUGCUUCU- ------..((((.((...)).))))((.((.((..(-((((((((((((.........)))))(.(((((((((......))))))))).)..))))-))))..))....)).)).- ( -29.30, z-score = -0.46, R) >droSim1.chr3L 7052183 108 + 22553184 ------UGGGGCGGCACUGCAGCGCGAUGUAGGAAU-CGGUGGAGCUGCAGUUGAAAAGCGGCUGUCAAUGAAAAUGAAAUUUUAUUGAGAUUUUUA-CCGAAACCCCCUGCUUCU- ------.(((((((..(((((......)))))...(-((((((((((((.........)))))(.(((((((((......))))))))).)..))))-))))......))))))).- ( -36.00, z-score = -1.95, R) >droWil1.scaffold_180698 5298396 99 - 11422946 AGACACAAAACUCAAGCAUUUGUGGAUGGAUGGAGUAUGGAGGAGAUGCAGUUGAAAUGCGGCUGUCAAUGAAAAUGAAAUUUUAUUGAGAUUUUUUUA------------------ .........((((...((((....))))....))))...((((((((.((((((.....))))))(((((((((......))))))))).)))))))).------------------ ( -19.50, z-score = -0.76, R) >droGri2.scaffold_15110 11073614 81 - 24565398 ----------------UGGUUUUGCUAUGCCUCA-U-CAGGCGAGCUGCAGUUGAAAAGCGGCUGUCAAUGAAAAUGAAAUUUUAUUGAGAUUUUUUUA------------------ ----------------.......(((.(((((..-.-.))))))))..((((((.....))))))(((((((((......)))))))))..........------------------ ( -20.00, z-score = -0.87, R) >consensus ______UGGGGCGGCACUGCAGCGCGAUGUUGGAAU_UGGUGGAGCUGCAGUUGAAAAGCGGCUGUCAAUGAAAAUGAAAUUUUAUUGAGAUUUUUA_CCGAAACCCCCUGCUUCU_ ......................................((.((((((((.........)))))).(((((((((......)))))))))...............)).))........ (-12.18 = -13.32 + 1.14)
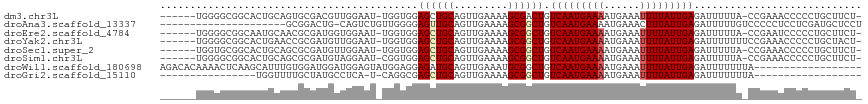
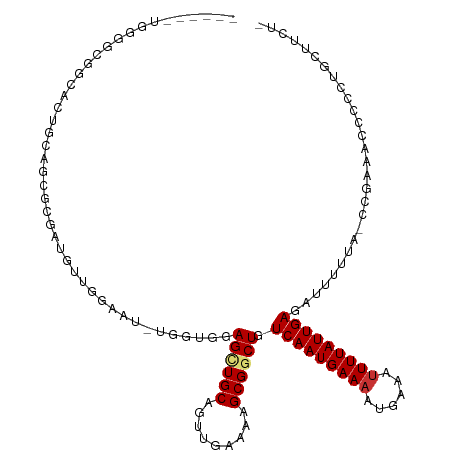

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:43 2011