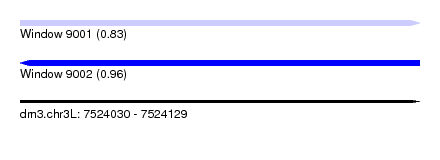
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,524,030 – 7,524,129 |
| Length | 99 |
| Max. P | 0.962267 |

| Location | 7,524,030 – 7,524,129 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.45 |
| Shannon entropy | 0.26500 |
| G+C content | 0.42762 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -12.89 |
| Energy contribution | -12.65 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
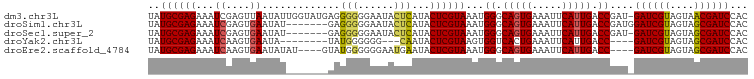
>dm3.chr3L 7524030 99 + 24543557 GUGGAUCGUUACUACGAUC-AUCGGUCAAUGAAUUUCACUGCCCAUUUACGAGUAUGAGUAUUCCCCCCUCAUACCAAUAUUAACUCGAUUUCUCGCAUA (((((((((....))))))-(((((.((.((.....)).)).))......(.(((((((.........))))))))...........)))....)))... ( -21.80, z-score = -2.41, R) >droSim1.chr3L 7038300 93 + 22553184 GUGGAUCGCUACUACGAUCCAUCGGUCAAUGAAUUUCACUGCCCAUUUACGAGUAUGAGUAUUCCCCCUC-------AUAUUCACUCGAUUUCUCGCAUA ((((((((......)))))))).((.((.((.....)).)).))......(((((((((........)))-------))))))................. ( -25.10, z-score = -3.61, R) >droSec1.super_2 7453519 92 + 7591821 GUGGAUCGCUACUACGAUC-AUCGGUCAAUGAAUUUCACUGCCCAUUUACGAGUAUGAGUAUUCCCCCUC-------AUAUUCACUCGAUUUCUCGCAUA ((((((((......)))))-(((((.((.((.....)).)).))......(((((((((........)))-------))))))....)))....)))... ( -23.60, z-score = -3.03, R) >droYak2.chr3L 8127689 85 + 24197627 GUGGAUCGCUACUACGAUC----GGUCAAUGAAUUUCAGUGACCACUUACGAGUAUUG---CCCCCCAUA--------UAUUCACUUGAUUUCUCGCAUA ((((((((......)))))----(((((.((.....)).)))))......((((((..---........)--------)))))...........)))... ( -19.70, z-score = -1.95, R) >droEre2.scaffold_4784 21947173 92 - 25762168 GUGGAUCGCUACUACGAUC----GGUCAAUGAAUUUCACUGCCCAUUUACGAGUAUUCAUUCCCCCCAUAC----AUAUAUUCACUUGAUUUCUCGCAUA ((((((((......)))))----((..((((((((((.............))).)))))))..))......----...................)))... ( -15.32, z-score = -1.07, R) >consensus GUGGAUCGCUACUACGAUC_AUCGGUCAAUGAAUUUCACUGCCCAUUUACGAGUAUGAGUAUUCCCCCUA_______AUAUUCACUCGAUUUCUCGCAUA ((((((((......)))))....((.((.((.....)).)).)).....(((((.............................)))))......)))... (-12.89 = -12.65 + -0.24)

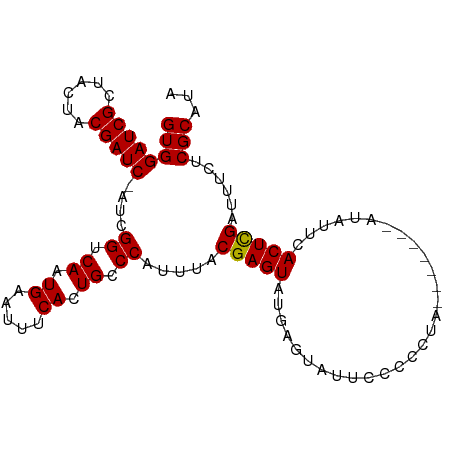
| Location | 7,524,030 – 7,524,129 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.45 |
| Shannon entropy | 0.26500 |
| G+C content | 0.42762 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7524030 99 - 24543557 UAUGCGAGAAAUCGAGUUAAUAUUGGUAUGAGGGGGGAAUACUCAUACUCGUAAAUGGGCAGUGAAAUUCAUUGACCGAU-GAUCGUAGUAACGAUCCAC ....(((....)))......(((.((((((((.........)))))))).))).((.(((((((.....))))).)).))-((((((....))))))... ( -28.90, z-score = -2.79, R) >droSim1.chr3L 7038300 93 - 22553184 UAUGCGAGAAAUCGAGUGAAUAU-------GAGGGGGAAUACUCAUACUCGUAAAUGGGCAGUGAAAUUCAUUGACCGAUGGAUCGUAGUAGCGAUCCAC ..((((((...((....)).(((-------(((........))))))))))))..(((.(((((.....))))).))).((((((((....)))))))). ( -31.60, z-score = -3.96, R) >droSec1.super_2 7453519 92 - 7591821 UAUGCGAGAAAUCGAGUGAAUAU-------GAGGGGGAAUACUCAUACUCGUAAAUGGGCAGUGAAAUUCAUUGACCGAU-GAUCGUAGUAGCGAUCCAC ..((((((...((....)).(((-------(((........)))))))))))).((.(((((((.....))))).)).))-((((((....))))))... ( -27.30, z-score = -2.66, R) >droYak2.chr3L 8127689 85 - 24197627 UAUGCGAGAAAUCAAGUGAAUA--------UAUGGGGGG---CAAUACUCGUAAGUGGUCACUGAAAUUCAUUGACC----GAUCGUAGUAGCGAUCCAC .....((....))..(((....--------((((((...---.....))))))...(((((.((.....)).)))))----((((((....))))))))) ( -23.30, z-score = -1.92, R) >droEre2.scaffold_4784 21947173 92 + 25762168 UAUGCGAGAAAUCAAGUGAAUAUAU----GUAUGGGGGGAAUGAAUACUCGUAAAUGGGCAGUGAAAUUCAUUGACC----GAUCGUAGUAGCGAUCCAC .....((....))..(((....(((----.((((((...........)))))).)))(((((((.....))))).))----((((((....))))))))) ( -21.40, z-score = -1.20, R) >consensus UAUGCGAGAAAUCGAGUGAAUAU_______GAGGGGGAAUACUCAUACUCGUAAAUGGGCAGUGAAAUUCAUUGACCGAU_GAUCGUAGUAGCGAUCCAC ..((((((...((....)).............(((......)))...))))))...((.(((((.....))))).))....((((((....))))))... (-18.96 = -18.88 + -0.08)
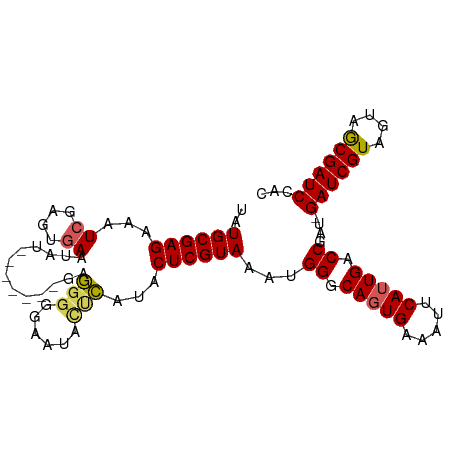
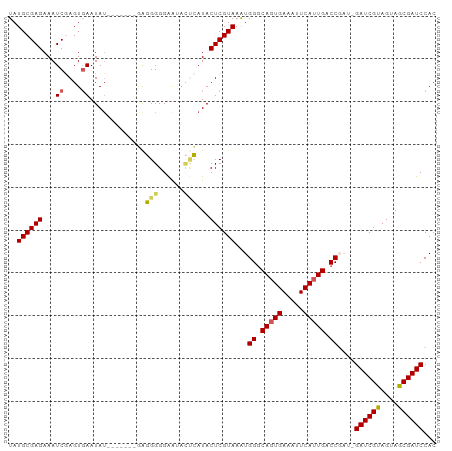
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:42 2011