| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,497,680 – 7,497,772 |
| Length | 92 |
| Max. P | 0.820000 |

| Location | 7,497,680 – 7,497,772 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.01 |
| Shannon entropy | 0.45683 |
| G+C content | 0.41292 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -9.76 |
| Energy contribution | -9.59 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
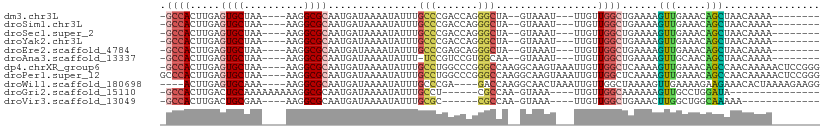
>dm3.chr3L 7497680 92 - 24543557 -GCCACUUGAGUGCUAA----AAGGCGCAAUGAUAAAAUAUUUGCCCGACCAGGGCUA--GUAAAU---UUGUUGGCUGAAAAGUUGAAACAGCUAACAAAA-------- -.........(((((..----..)))))..........((((.((((.....)))).)--)))..(---((((((((((...........))))))))))).-------- ( -28.40, z-score = -3.20, R) >droSim1.chr3L 7012668 92 - 22553184 -GCCACUUGAGUGCUAA----AAGGCGCAAUGAUAAAAUAUUUGCCCGACCAGGGCUA--GUAAAU---UUGUUGGCUGAAAAGUUGAAACAGCUAACAAAA-------- -.........(((((..----..)))))..........((((.((((.....)))).)--)))..(---((((((((((...........))))))))))).-------- ( -28.40, z-score = -3.20, R) >droSec1.super_2 7422860 92 - 7591821 -GCCACUUGAGUGCUAA----AAGGCGCAAUGAUAAAAUAUUUGCCCGACCAGGGCUA--GUAAAU---UUGUUGGCUGAAAAGUUGAAACAGCUAACAAAA-------- -.........(((((..----..)))))..........((((.((((.....)))).)--)))..(---((((((((((...........))))))))))).-------- ( -28.40, z-score = -3.20, R) >droYak2.chr3L 8100802 92 - 24197627 -GCCACUUGAGUGCUAA----AAGGCGCAAUGAUAAAAUAUUUGCCCGACCAGGGCUA--GUAAAU---UUGUUGGCUGAAAAGUUGAAACAGCUAACAAAA-------- -.........(((((..----..)))))..........((((.((((.....)))).)--)))..(---((((((((((...........))))))))))).-------- ( -28.40, z-score = -3.20, R) >droEre2.scaffold_4784 21920637 92 + 25762168 -GCCACUUGAGUGCUAA----AAGGCGCAAUGAUAAAAUAUUUGCCCGAGCAGGGCUA--GUAAAU---UUGUUGGCUGAAAAGUUGAAACAGCUAACAAAA-------- -.........(((((..----..)))))..........((((.((((.....)))).)--)))..(---((((((((((...........))))))))))).-------- ( -28.40, z-score = -2.84, R) >droAna3.scaffold_13337 16282160 91 - 23293914 -GCCACUUGAGUGCUAA----AAGGCGCAAUGAUAAAAUAUUU-UCCGUCCGUGGCAA--GUAAAU---UUGUUGGCUGAAAAGUUGCAACAGCUAACAAAA-------- -(((((....(((((..----..)))))...(((..((....)-)..))).)))))..--.....(---((((((((((...........))))))))))).-------- ( -27.30, z-score = -2.82, R) >dp4.chrXR_group6 1495062 105 - 13314419 -GCCACUUGAGUGCUAA----AAGGCGCAAUGAUAAAAUAUUUGCCUGGCCCGGGCCAAGGCAAGUAAAUUGUUGGCUCAAAAGUUGAAACAGCCAACAAAAACUCCGGG -.((......(((((..----..)))))..........((((((((((((....))).)))))))))..(((((((((.............))))))))).......)). ( -36.92, z-score = -2.97, R) >droPer1.super_12 2184819 106 - 2414086 GCCCACUUGAGUGCUAA----AAGGCGCAAUGAUAAAAUAUUUGCCUGGCCCGGGCCAAGGCAAGUAAAUUGUUGGCUCAAAAGUUGAAACAGCCAACAAAAACUCCGGG .(((......(((((..----..)))))..........((((((((((((....))).)))))))))..(((((((((.............))))))))).......))) ( -38.92, z-score = -3.41, R) >droWil1.scaffold_180698 6036844 98 - 11422946 ----ACUUGAGUGCAAA----AAGGCGCAAUGAUAAAAUAUUUGCCCGA----GACCAAGGCAACUAAAUUGUUGGCUAAAAGUUGAAAAGAAGAAACACUAAAAGAAGG ----.(((.((((....----..((.((((...........))))))..----.....((.((((......)))).))...................))))..))).... ( -16.20, z-score = -0.54, R) >droGri2.scaffold_15110 11030720 83 + 24565398 -GCCACUUGACUGCAAAAAAAAAGGCGCAAUGAUAAAAUAUUUGCCU------CGCCAA-GUAAA----UUGUUGGCAAAAAAGUUGCCUGGAUA--------------- -.(((..(((((..........(((((.((((......)))))))))------.(((((-.....----...))))).....)))))..)))...--------------- ( -17.00, z-score = -0.29, R) >droVir3.scaffold_13049 5485601 81 - 25233164 -GCCACUUGACUGCGAA----AAGGCGCAAUGAUAAAAUAUUUGCGC------CGCCAA-GUAAA----UUGUUGGCUGAAACUUGGCUGGCAAAAA------------- -((((............----..(((((((...........))))))------)(((((-((...----..(....)....))))))))))).....------------- ( -27.10, z-score = -2.91, R) >consensus _GCCACUUGAGUGCUAA____AAGGCGCAAUGAUAAAAUAUUUGCCCGACCAGGGCCA__GUAAAU___UUGUUGGCUGAAAAGUUGAAACAGCUAACAAAA________ .(((......((((..........))))...(((((...((((((...............))))))...))))))))......(((.....)))................ ( -9.76 = -9.59 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:40 2011