| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,457,751 – 7,457,857 |
| Length | 106 |
| Max. P | 0.505899 |

| Location | 7,457,751 – 7,457,857 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.41233 |
| G+C content | 0.46652 |
| Mean single sequence MFE | -24.39 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.71 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7457751 106 + 24543557 GAAUUUCCUGCUACUGCCACUGCUUUGGAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCACGAGCGUGCACAUGACACCAGCCCACGCACA--UUUAUCAGCCG----- .........((((((.(((......)))))).((((((((((((((.....))))))))........(((((....((....))...)))))...--)))))))))..----- ( -24.10, z-score = -0.89, R) >droAna3.scaffold_13337 16247658 107 + 23293914 ---GUACAUGCCUUGGCCUCUGCUG-UGAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCACGAGCGUGCACUUGACACCCGCCCACGCACA--UUUAUCAGCCCGGGGA ---.......(((.((((((.....-.)))..(((((((..(((((((..((.....))...)))))))((((.................)))))--)))))).))).))).. ( -28.33, z-score = -0.22, R) >droEre2.scaffold_4784 21881624 106 - 25762168 GAAUGCGUUGCUACUGCCAGUGCUUGGGAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCACGAGCGUGCACAUGACACCAGCCCACGCACA--UUUAUCAGCCG----- ....((...((....))..((((.((((.((((.....((((((((.....))))))))...(((....)))....)))).....)))).)))).--.......))..----- ( -26.50, z-score = 0.03, R) >droYak2.chr3L 8062894 98 + 24197627 ------GAAUACGCUGGAGUCGUU--GGAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCACGAGCGUGCACAUGACACCAGCCCACGCACA--UUUAUCAGCCG----- ------......(((.((.((...--.)).))((((((((((((((.....))))))))........(((((....((....))...)))))...--)))))))))..----- ( -24.40, z-score = -0.71, R) >droSec1.super_2 7388903 106 + 7591821 GAAUUUGCUUCAACUGCCACUGCUUUGGAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCAUGAGCGUGCACAUGACACCAGCCCACGCACA--UUUAUCAGCCG----- ......(((((((..((....)).))))))).((((((((((((((.....)))))))).....((.(((((....((....))...))))).))--)))))).....----- ( -25.90, z-score = -1.16, R) >droSim1.chr3L 6978657 106 + 22553184 GAAUUUGCUUCAACUGACACUGCUUUGGAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCACGAGCGUGCACAUGACACCAGCCCACGCACA--UUUAUCAGCCG----- ......(((......(((.((.....)).)))((((((((((((((.....))))))))........(((((....((....))...)))))...--)))))))))..----- ( -23.70, z-score = -0.58, R) >droMoj3.scaffold_6680 3796791 101 + 24764193 -----CUACUACAGCU-CUCUGCUC-AGCAUCGAUAAAUCAGUUUGUAUAUCAAAUUGAAUUCACGAGCGUGCACUUGACACCAGCACACGCACA--UUUAUCAGCCCAG--- -----........(((-........-)))...((((((((((((((.....))))))))........(((((...(((....)))..)))))...--)))))).......--- ( -20.80, z-score = -1.60, R) >droVir3.scaffold_13049 5441960 104 + 25233164 -GGGUCUCCGGCUCCGGCUCUGCUC-GGCAUCGAUAAAUCAGUUUGUAUAUCAAAUUGAAUUCACGAGCGUGCACUUGACACCAGCACACGCACA--UUUAUCAGCCG----- -((((..(((....)))....))))-(((...((((((((((((((.....))))))))........(((((...(((....)))..)))))...--)))))).))).----- ( -30.00, z-score = -2.05, R) >droGri2.scaffold_15110 10995194 94 - 24565398 --------------UUCCUCUACUU-CACAUCAAUAAAUCAGUUUGUAUAUCAAAUUGAAUUCACGAGCGUGCACUUGACACCAGCACACGCACACAUUUAUCAGCCGG---- --------------..((.......-............((((((((.....))))))))........(((((...(((....)))..)))))...............))---- ( -14.80, z-score = -1.49, R) >dp4.chrXR_group6 1457873 94 + 13314419 --------UGGCACUGCCACAGC----GAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCACGAGCGUGCACUUGACACCCGCACACGCACA--UUUAUCAGCCA----- --------.((((((((....))----.))).(((((((..(((((((..((.....))...)))))))((((...........))))......)--)))))).))).----- ( -24.90, z-score = -1.38, R) >droPer1.super_12 2147396 94 + 2414086 --------UGGCACUGCCACAGC----GAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCACGAGCGUGCACUUGACACCCGCACACGCACA--UUUAUCAGCCA----- --------.((((((((....))----.))).(((((((..(((((((..((.....))...)))))))((((...........))))......)--)))))).))).----- ( -24.90, z-score = -1.38, R) >consensus ______GCUGCCACUGCCACUGCUU_GGAGUCGAUAAAUCAGUUUGUGUAUCAAAUUGAAUUCACGAGCGUGCACUUGACACCAGCCCACGCACA__UUUAUCAGCCG_____ ................................((((((((((((((.....))))))))........(((((..(.........)..))))).....)))))).......... (-16.62 = -16.71 + 0.09)

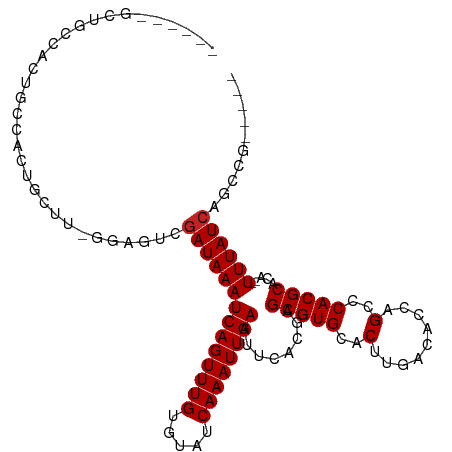

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:38 2011