
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,452,278 – 7,452,385 |
| Length | 107 |
| Max. P | 0.519278 |

| Location | 7,452,278 – 7,452,385 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Shannon entropy | 0.44133 |
| G+C content | 0.35939 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -13.93 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7452278 107 + 24543557 AUUACGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAUGC--CUAAUAAAAUUUUCAUGC-----CCACACACGACGGAGUGCAUCGCA----- ((((.((((....((((..(((((((((((..-(((.....)))...))))))))))))))).)))--)))))..........(((-----.....(((......)))....)))----- ( -22.70, z-score = -0.05, R) >droSim1.chr3L 6973337 107 + 22553184 AUUACGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAUGC--CUAAUAAAAUUUUCAUGC-----CCACACACGGCGGAGUGCAUCGCA----- ....(((((.....)))))(((((((((((..-(((.....)))...)))))))))))((..((((--((.............(((-----(.......)))).)).)))).)).----- ( -24.84, z-score = -0.30, R) >droSec1.super_2 7383558 97 + 7591821 AUUACGGCAACAAGUG----------UCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAUGC--CUAAUAAAAUUUUCAUGC-----CCACACACGGCGGAGUGCAUCGCA----- ......((.....(((----------((((..-(((.....)))...)))))))...(((((.(((--(.................-----........))))..)))))..)).----- ( -19.31, z-score = 0.27, R) >droYak2.chr3L 8057157 112 + 24197627 AUUACGGCAACAAGUGUCGAAAUGUGUCGAAAAUAUUGCAAAUAAAGUUGACACAUUUGCAUAUGC--CUAAUAAAAUUUUCAUACAUUUUCCGCACACGGCG-AGUGCAGCGCA----- ((((.((((....((((..(((((((((((...(((.....)))...))))))))))))))).)))--)))))............(((((.(((....))).)-)))).......----- ( -25.90, z-score = -0.60, R) >droEre2.scaffold_4784 21873294 106 - 25762168 AUUACGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAUGC--CUAAUAAAAUUUUCAUAC-----CCAAACACGGCG-AGUGCACCGCA----- ....(((((.....)))))(((((((((((..-(((.....)))...)))))))))))((((.(((--(.................-----........))))-.))))......----- ( -25.41, z-score = -1.57, R) >droAna3.scaffold_13337 16242505 99 + 23293914 AUUAUGGCAACAAGUGUCGAAAUGUGUCGAAA-UGUUGCAAAUAAAAUUGACACAUUUGCAUAUAAGCCUAAUAAAAUUUUUAUUG-------GCAUGUGUUGGAGC------------- .......(((((.((((..(((((((((((..-(((.....)))...)))))))))))))))....(((.(((((.....))))))-------))...)))))....------------- ( -23.20, z-score = -1.39, R) >dp4.chrXR_group6 1452192 104 + 13314419 AUUAUGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAC----CUCAUAAAAUUU-C-CAU-------GCACAUGUGCAAAAGUUUCAUAACG-- .((((((.(((..((((..(((((((((((..-(((.....)))...)))))))))))..))))----............-.-..(-------(((....))))...)))))))))..-- ( -23.20, z-score = -1.61, R) >droPer1.super_12 2141714 104 + 2414086 AUUAUGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAC----CUCAUAAAAUUU-C-CAU-------GCACAUGUGCAAAAGUUUCAUAACG-- .((((((.(((..((((..(((((((((((..-(((.....)))...)))))))))))..))))----............-.-..(-------(((....))))...)))))))))..-- ( -23.20, z-score = -1.61, R) >droWil1.scaffold_180698 5985936 105 + 11422946 AUUAUGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAC----CUAAUAAAAUUUUCAUGC-------ACACAUGUAUGGGAAAUCCACGCA--- ....((....)).((((.((((((((((((..-(((.....)))...)))))))))).......----.........((..(((((-------(....))))))..)).)).)))).--- ( -25.70, z-score = -1.84, R) >droVir3.scaffold_13049 5435255 98 + 25233164 AUUAUGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAC----CUAAUAAAAUUUUC-CAU-------GCACAUGUGGAAA-GUUAC-------- ....((....)).((((..(((((((((((..-(((.....)))...)))))))))))..))))----.......(((((((-(((-------......)))))))-)))..-------- ( -25.70, z-score = -2.89, R) >droMoj3.scaffold_6680 3790312 101 + 24764193 AUUAUGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAGUUGACACAUUUGCAUAC----CUAAUAAAAUUUUC-CAU-------GCACAUGCGGAAAAGUUGUGA------ ......(((((..((((..(((((((((((..-(((.....)))...)))))))))))..))))----.........(((((-(..-------((....)))))))))))))..------ ( -25.40, z-score = -2.06, R) >droGri2.scaffold_15110 10990141 102 - 24565398 AUUAUGGCAACAAGUGUCGAAAUGUGUCGAAA-UAUUGCAAAUAAAAUUGACACAUUUGCAUACUA--CUAAUAAAAUG-----AU-------GCAUGUG---GAAAACUGGAUGCCGCU .....((((.(.(((.((((((((((((((..-(((.....)))...)))))))))))((((.(..--..........)-----))-------))...))---)...))).).))))... ( -22.30, z-score = -1.48, R) >consensus AUUAUGGCAACAAGUGUCGAAAUGUGUCGAAA_UAUUGCAAAUAAAGUUGACACAUUUGCAUAC____CUAAUAAAAUUUUCAUAC_______GCACAUGUCGGAGAGCUUCGCA_____ .....(....)..((((..(((((((((((...(((.....)))...))))))))))))))).......................................................... (-13.93 = -14.52 + 0.59)

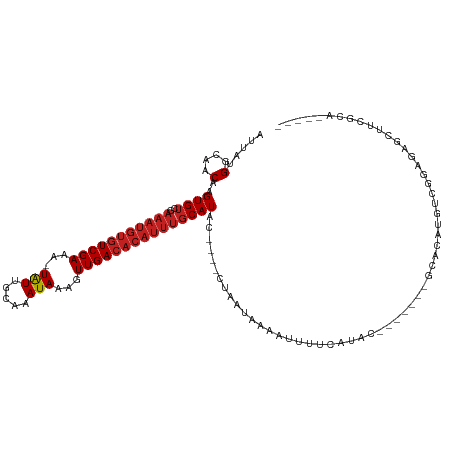
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:36 2011