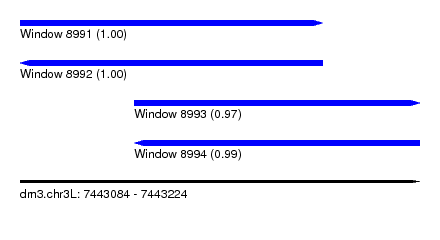
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,443,084 – 7,443,224 |
| Length | 140 |
| Max. P | 0.999596 |

| Location | 7,443,084 – 7,443,190 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.91 |
| Shannon entropy | 0.35553 |
| G+C content | 0.59944 |
| Mean single sequence MFE | -59.60 |
| Consensus MFE | -36.58 |
| Energy contribution | -36.44 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.23 |
| Mean z-score | -4.31 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
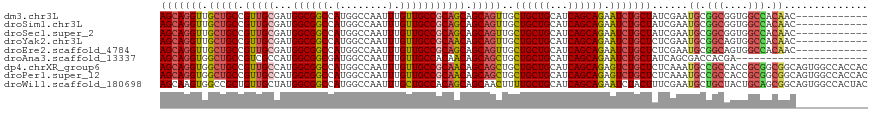
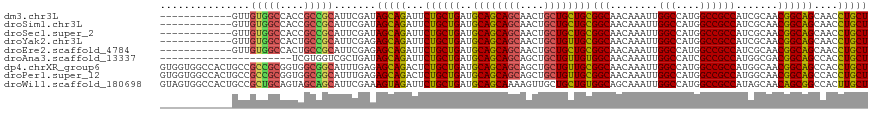
>dm3.chr3L 7443084 106 + 24543557 AGCAGGUUGCUGCCGUUGCGAUGGCGGCCAUGGCCAAUUUGUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGAAUCUGCUAUCGAAUGCGGCGGUGGCCACAAC------------ ....(((..(((((((..((((((((((....)))..(((((((..((((((((....))))))))..)))))))....)))))))...)))))))..))).....------------ ( -64.10, z-score = -5.92, R) >droSim1.chr3L 6964142 106 + 22553184 AGCAGGUUGCUGCCGUUGCGAUGGCGGCCAUGGCCAAUUUGUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGAAUCUGCUAUCGAAUGCGGCGGUGGCCACAAC------------ ....(((..(((((((..((((((((((....)))..(((((((..((((((((....))))))))..)))))))....)))))))...)))))))..))).....------------ ( -64.10, z-score = -5.92, R) >droSec1.super_2 7374397 106 + 7591821 AGCAGGUUGCUGCCGUUGCGAUGGCGGCCAUGGCCAAUUUGUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGAAUCUGCUAUCGAAUGCGGCGGUGGCCACAAC------------ ....(((..(((((((..((((((((((....)))..(((((((..((((((((....))))))))..)))))))....)))))))...)))))))..))).....------------ ( -64.10, z-score = -5.92, R) >droYak2.chr3L 8047918 106 + 24197627 AGCAGGUUGCUGCCGUUGCGAUGGCGGCCAUGGCCAAUUUGUUGCCGCAACAGCAGUUGCUGCUGCAUCAGCAGAAUCUGCUCUCGAAUGCGGCAGUGGCCACAAC------------ ....(((..(((((((..(((.((((((....)))..(((((((..(((.((((....)))).)))..)))))))....))).)))...)))))))..))).....------------ ( -54.40, z-score = -3.65, R) >droEre2.scaffold_4784 21864157 106 - 25762168 AGCAGGUUGCUGCCGUUGCGAUGGCGGCCAUGGCCAAUUUGUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGAAUCUGCUCUCGAAUGCGGCAGUGGCCACAAC------------ ....(((..(((((((..(((.((((((....)))..(((((((..((((((((....))))))))..)))))))....))).)))...)))))))..))).....------------ ( -61.00, z-score = -5.01, R) >droAna3.scaffold_13337 16233154 96 + 23293914 AGCAGGUGGCUGCCGUCGCCAUGGCGGCGAUGGCCAAUUUGUUGCCACAACAGCAGCUGCUGCUGCAUCAGCAGAAUCUGCUAUCAGCGACCACGA---------------------- ....(((.((((((((((((.....)))))))))......((((...))))(((((...((((((...))))))...)))))...))).)))....---------------------- ( -43.70, z-score = -2.13, R) >dp4.chrXR_group6 1441279 118 + 13314419 AGCAGGUGGCUGCCGUUGCCAUGGCGGCCAUGGCCAAUUUGUUGCCGCAACAGCAGCUGCUGCUGCAUCAGCAGAGUCUGCUCUCAAAUGCCGCCACCGCGGCGGCAGUGGCCACCAC ....(((.(((((((((((..(((((((...(((.........))).....(((((((.((((((...)))))))).))))).......)))))))..))))))))))).)))..... ( -62.60, z-score = -2.79, R) >droPer1.super_12 2131003 118 + 2414086 AGCAGGUGGCUGCCGUUGCCAUGGCGGCCAUGGCCAAUUUGUUGCCGCAACAGCAGCUGCUGCUGCAUCAGCAGAGUCUGCUCUCAAAUGCCGCCACCGCGGCGGCAGUGGCCACCAC ....(((.(((((((((((..(((((((...(((.........))).....(((((((.((((((...)))))))).))))).......)))))))..))))))))))).)))..... ( -62.60, z-score = -2.79, R) >droWil1.scaffold_180698 5972610 118 + 11422946 AGCAAGUGGCCGCUGUUGCUAUGGCGGCCAUGGCCAAUUUGCUGCCACAGCAGCAACUUUUGCUGCAUCAGCAGAAUCUACUUUCGAAUGCUGCUACUGCAGCGGCAGUGGCCACUAC ....((((((((((((((((..((((((....)))...(((((((....))))))).....)))(((..(((((..((.......))...)))))..))))))))))))))))))).. ( -59.80, z-score = -4.64, R) >consensus AGCAGGUUGCUGCCGUUGCGAUGGCGGCCAUGGCCAAUUUGUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGAAUCUGCUAUCGAAUGCGGCGGUGGCCACAAC____________ (((((((.(((((.(((((...((((((.(........).))))))))))).)))))..((((((...)))))).)))))))......((.(((....))).)).............. (-36.58 = -36.44 + -0.13)
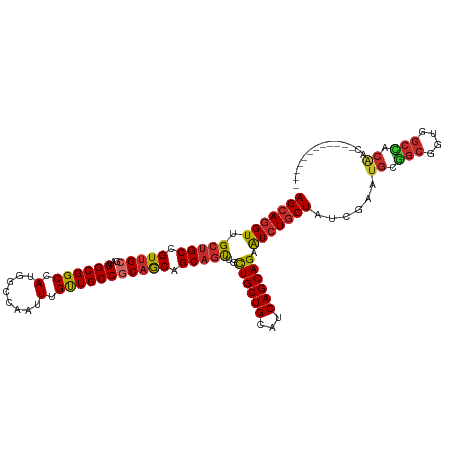
| Location | 7,443,084 – 7,443,190 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.91 |
| Shannon entropy | 0.35553 |
| G+C content | 0.59944 |
| Mean single sequence MFE | -52.94 |
| Consensus MFE | -43.76 |
| Energy contribution | -43.12 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
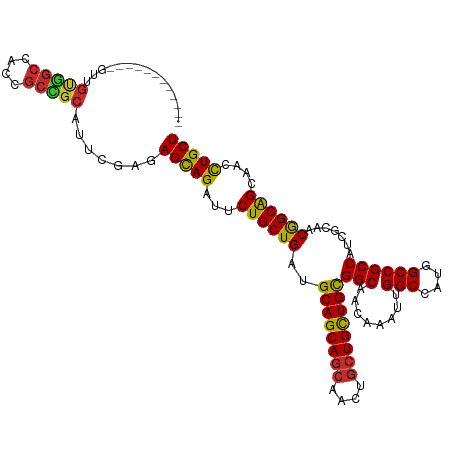
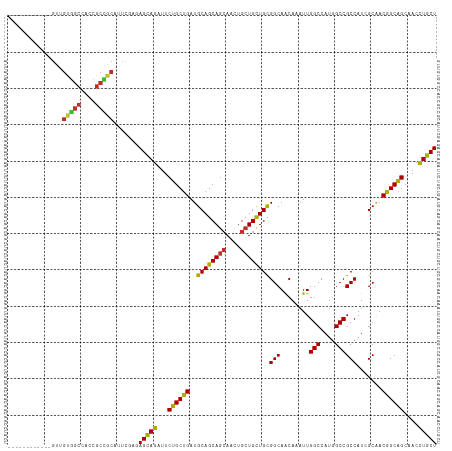
>dm3.chr3L 7443084 106 - 24543557 ------------GUUGUGGCCACCGCCGCAUUCGAUAGCAGAUUCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAACAAAUUGGCCAUGGCCGCCAUCGCAACGGCAGCAACCUGCU ------------..((((((....))))))......(((((.((((((((..((((((((....))))))))(((........(((....)))))).......)))))).)).))))) ( -48.70, z-score = -2.75, R) >droSim1.chr3L 6964142 106 - 22553184 ------------GUUGUGGCCACCGCCGCAUUCGAUAGCAGAUUCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAACAAAUUGGCCAUGGCCGCCAUCGCAACGGCAGCAACCUGCU ------------..((((((....))))))......(((((.((((((((..((((((((....))))))))(((........(((....)))))).......)))))).)).))))) ( -48.70, z-score = -2.75, R) >droSec1.super_2 7374397 106 - 7591821 ------------GUUGUGGCCACCGCCGCAUUCGAUAGCAGAUUCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAACAAAUUGGCCAUGGCCGCCAUCGCAACGGCAGCAACCUGCU ------------..((((((....))))))......(((((.((((((((..((((((((....))))))))(((........(((....)))))).......)))))).)).))))) ( -48.70, z-score = -2.75, R) >droYak2.chr3L 8047918 106 - 24197627 ------------GUUGUGGCCACUGCCGCAUUCGAGAGCAGAUUCUGCUGAUGCAGCAGCAACUGCUGUUGCGGCAACAAAUUGGCCAUGGCCGCCAUCGCAACGGCAGCAACCUGCU ------------..((((((....))))))......(((((.((((((((..((((((((....))))))))(((........(((....)))))).......)))))).)).))))) ( -46.40, z-score = -1.93, R) >droEre2.scaffold_4784 21864157 106 + 25762168 ------------GUUGUGGCCACUGCCGCAUUCGAGAGCAGAUUCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAACAAAUUGGCCAUGGCCGCCAUCGCAACGGCAGCAACCUGCU ------------..((((((....))))))......(((((.((((((((..((((((((....))))))))(((........(((....)))))).......)))))).)).))))) ( -48.80, z-score = -2.41, R) >droAna3.scaffold_13337 16233154 96 - 23293914 ----------------------UCGUGGUCGCUGAUAGCAGAUUCUGCUGAUGCAGCAGCAGCUGCUGUUGUGGCAACAAAUUGGCCAUCGCCGCCAUGGCGACGGCAGCCACCUGCU ----------------------..(((((.(((((((((((...((((((...))))))...))))))))..))).........(((.(((((.....))))).))).)))))..... ( -43.10, z-score = -1.70, R) >dp4.chrXR_group6 1441279 118 - 13314419 GUGGUGGCCACUGCCGCCGCGGUGGCGGCAUUUGAGAGCAGACUCUGCUGAUGCAGCAGCAGCUGCUGUUGCGGCAACAAAUUGGCCAUGGCCGCCAUGGCAACGGCAGCCACCUGCU ((((((((...(((((..((.((((((((...((((((....))))((..((((((((((....))))))))(....)..))..))))..)))))))).))..))))))))))).)). ( -65.80, z-score = -3.14, R) >droPer1.super_12 2131003 118 - 2414086 GUGGUGGCCACUGCCGCCGCGGUGGCGGCAUUUGAGAGCAGACUCUGCUGAUGCAGCAGCAGCUGCUGUUGCGGCAACAAAUUGGCCAUGGCCGCCAUGGCAACGGCAGCCACCUGCU ((((((((...(((((..((.((((((((...((((((....))))((..((((((((((....))))))))(....)..))..))))..)))))))).))..))))))))))).)). ( -65.80, z-score = -3.14, R) >droWil1.scaffold_180698 5972610 118 - 11422946 GUAGUGGCCACUGCCGCUGCAGUAGCAGCAUUCGAAAGUAGAUUCUGCUGAUGCAGCAAAAGUUGCUGCUGUGGCAGCAAAUUGGCCAUGGCCGCCAUAGCAACAGCGGCCACUUGCU (((((((((......((((((.((((((.((((....)..))).)))))).))))))....((((.(((((((((........(((....)))))))))))).)))))))))).))). ( -60.50, z-score = -4.29, R) >consensus ____________GUUGUGGCCACCGCCGCAUUCGAGAGCAGAUUCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAACAAAUUGGCCAUGGCCGCCAUCGCAACGGCAGCAACCUGCU ...............(((((....))))).......(((((...((((((..((((((((....))))))))(((........(((....)))))).......))))))....))))) (-43.76 = -43.12 + -0.64)
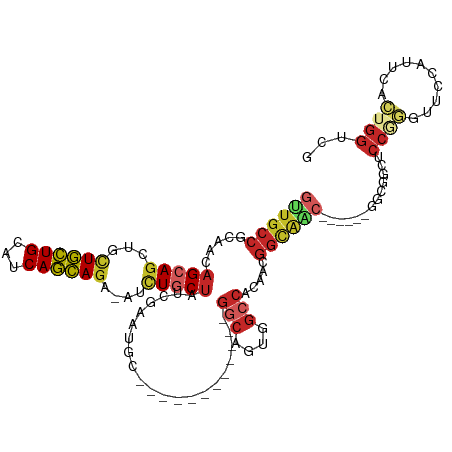
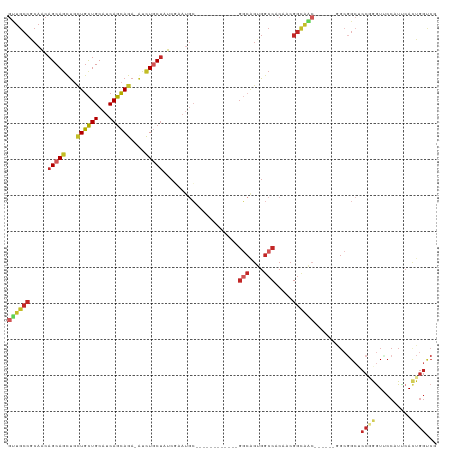
| Location | 7,443,124 – 7,443,224 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.63 |
| Shannon entropy | 0.57205 |
| G+C content | 0.61028 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -20.95 |
| Energy contribution | -22.08 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967753 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
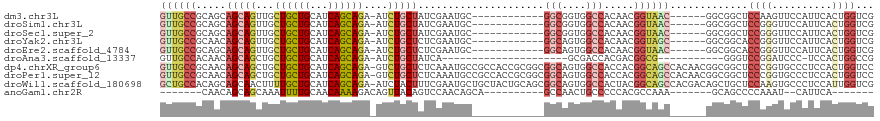
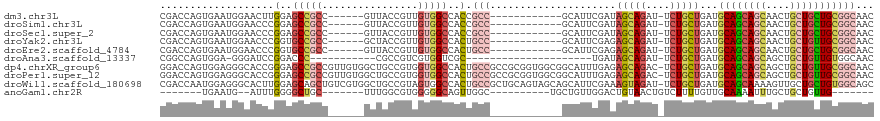
>dm3.chr3L 7443124 100 + 24543557 GUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGA-AUCUGCUAUCGAAUGC------------GGCGGUGGCCACAACGGUAAC------GGCGGCUCCAAGUUCCAUUCACUGGUCG ((((((((((((((....))))))))....((((.-..))))(((((..((.------------(((....))).)).)))))..------))))))........(((.....)))... ( -39.20, z-score = -1.41, R) >droSim1.chr3L 6964182 100 + 22553184 GUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGA-AUCUGCUAUCGAAUGC------------GGCGGUGGCCACAACGGUAAC------GGCGGCUCCGGGUUCCAUUCACUGGUCG ((((((((((((((....))))))))....((((.-..))))(((((..((.------------(((....))).)).)))))..------)))))).((((..........))))... ( -41.80, z-score = -1.61, R) >droSec1.super_2 7374437 100 + 7591821 GUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGA-AUCUGCUAUCGAAUGC------------GGCGGUGGCCACAACGGUAAC------GGCGGCUCCGGGUUCCAUUCACUGGUCG ((((((((((((((....))))))))....((((.-..))))(((((..((.------------(((....))).)).)))))..------)))))).((((..........))))... ( -41.80, z-score = -1.61, R) >droYak2.chr3L 8047958 100 + 24197627 GUUGCCGCAACAGCAGUUGCUGCUGCAUCAGCAGA-AUCUGCUCUCGAAUGC------------GGCAGUGGCCACAACGGUAGC------GGCGGCACCGGGUUCCAUUCACUGGUCG ((((((((.((....((..(((((((((((((((.-..)))))...).))))------------)))))..)).......)).))------))))))(((((..........))))).. ( -44.50, z-score = -2.52, R) >droEre2.scaffold_4784 21864197 100 - 25762168 GUUGCCGCAGCAGCAGUUGCUGCUGCAUCAGCAGA-AUCUGCUCUCGAAUGC------------GGCAGUGGCCACAACGGUAAC------GGCGGCACCGGGUUCCAUUCACUGGUCG ((((((((((((((....))))))))((((((((.-..)))))......((.------------(((....))).))..)))...------))))))(((((..........))))).. ( -42.10, z-score = -1.79, R) >droAna3.scaffold_13337 16233194 85 + 23293914 GUUGCCACAACAGCAGCUGCUGCUGCAUCAGCAGA-AUCUGCUAUCA---------------------GCGACCACGACGGCG-----------GGGUCCGGAUCCC-UCCACUGGCCG ...((((....(((((...((((((...)))))).-..)))))....---------------------...........((.(-----------((((....)))))-.))..)))).. ( -30.90, z-score = -0.96, R) >dp4.chrXR_group6 1441319 118 + 13314419 GUUGCCGCAACAGCAGCUGCUGCUGCAUCAGCAGA-GUCUGCUCUCAAAUGCCGCCACCGCGGCGGCAGUGGCCACCACGGCAGCCACAACGGCGGCUCCCGGUGCCCUCCACUGGUCC (((((((....(((((((.((((((...)))))))-).)))))......(((((((.....)))))))(((((..(....)..)))))..)))))))..((((((.....))))))... ( -57.30, z-score = -2.48, R) >droPer1.super_12 2131043 118 + 2414086 GUUGCCGCAACAGCAGCUGCUGCUGCAUCAGCAGA-GUCUGCUCUCAAAUGCCGCCACCGCGGCGGCAGUGGCCACCACGGCAGCCACAACGGCGGCUCCCGGUGCCCUCCACUGGUCC (((((((....(((((((.((((((...)))))))-).)))))......(((((((.....)))))))(((((..(....)..)))))..)))))))..((((((.....))))))... ( -57.30, z-score = -2.48, R) >droWil1.scaffold_180698 5972650 118 + 11422946 GCUGCCACAGCAGCAACUUUUGCUGCAUCAGCAGA-AUCUACUUUCGAAUGCUGCUACUGCAGCGGCAGUGGCCACUACGGCAGCCACGACAGCUGCUCCAAGUGCCCUCCAUUGGUCG (((((...(((((((..((((((((...)))))))-)((.......)).)))))))...)))))((.((.(((.(((..((((((.......))))))...))))))))))........ ( -46.00, z-score = -2.16, R) >anoGam1.chr2R 34183486 86 - 62725911 -------CAACAGCAGCAAAUUUUGCAACAAAAGACAGUUACAGUCCAACAGCA----------GCCAACUGCCCCCACGCCAAA-------GCAGCCCCAAAU--CAUUCA------- -------.....((.((...(((.((.......(((.......))).....(((----------(....))))......)).)))-------)).)).......--......------- ( -9.80, z-score = -0.53, R) >consensus GUUGCCGCAACAGCAGCUGCUGCUGCAUCAGCAGA_AUCUGCUAUCGAAUGC____________GGCAGUGGCCACAACGGCAAC______GGCGGCUCCGGGUUCCAUUCACUGGUCG ((((((.....(((((...((((((...))))))....))))).....................(((....))).....)))))).............((((..........))))... (-20.95 = -22.08 + 1.13)
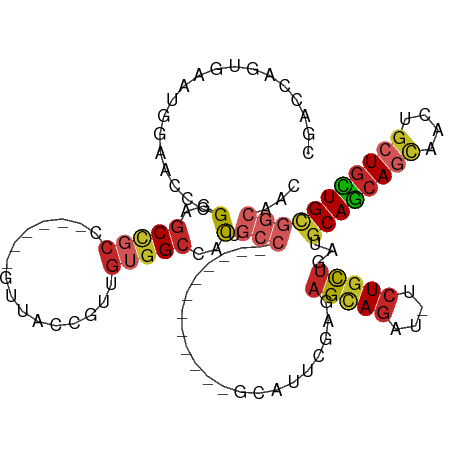
| Location | 7,443,124 – 7,443,224 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.63 |
| Shannon entropy | 0.57205 |
| G+C content | 0.61028 |
| Mean single sequence MFE | -44.34 |
| Consensus MFE | -23.41 |
| Energy contribution | -24.41 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986186 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 7443124 100 - 24543557 CGACCAGUGAAUGGAACUUGGAGCCGCC------GUUACCGUUGUGGCCACCGCC------------GCAUUCGAUAGCAGAU-UCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAAC ...(((((.......)).)))....(((------.....((.((((((....)))------------)))..)).((((((..-.))))))..((((((((....)))))))))))... ( -40.60, z-score = -2.49, R) >droSim1.chr3L 6964182 100 - 22553184 CGACCAGUGAAUGGAACCCGGAGCCGCC------GUUACCGUUGUGGCCACCGCC------------GCAUUCGAUAGCAGAU-UCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAAC ...(((.....)))...........(((------.....((.((((((....)))------------)))..)).((((((..-.))))))..((((((((....)))))))))))... ( -40.10, z-score = -2.34, R) >droSec1.super_2 7374437 100 - 7591821 CGACCAGUGAAUGGAACCCGGAGCCGCC------GUUACCGUUGUGGCCACCGCC------------GCAUUCGAUAGCAGAU-UCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAAC ...(((.....)))...........(((------.....((.((((((....)))------------)))..)).((((((..-.))))))..((((((((....)))))))))))... ( -40.10, z-score = -2.34, R) >droYak2.chr3L 8047958 100 - 24197627 CGACCAGUGAAUGGAACCCGGUGCCGCC------GCUACCGUUGUGGCCACUGCC------------GCAUUCGAGAGCAGAU-UCUGCUGAUGCAGCAGCAACUGCUGUUGCGGCAAC ...(((.....))).......((((((.------((...((.((((((....)))------------)))..))..(((((..-.((((((...))))))...))))))).)))))).. ( -39.70, z-score = -1.67, R) >droEre2.scaffold_4784 21864197 100 + 25762168 CGACCAGUGAAUGGAACCCGGUGCCGCC------GUUACCGUUGUGGCCACUGCC------------GCAUUCGAGAGCAGAU-UCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAAC ...(((.....))).......((((...------.....((.((((((....)))------------)))..))..(((((..-.)))))...((((((((....)))))))))))).. ( -39.60, z-score = -1.64, R) >droAna3.scaffold_13337 16233194 85 - 23293914 CGGCCAGUGGA-GGGAUCCGGACCC-----------CGCCGUCGUGGUCGC---------------------UGAUAGCAGAU-UCUGCUGAUGCAGCAGCAGCUGCUGUUGUGGCAAC .(((((((((.-(((........))-----------).))).).)))))((---------------------(((((((((..-.((((((...))))))...))))))))..)))... ( -38.00, z-score = -1.68, R) >dp4.chrXR_group6 1441319 118 - 13314419 GGACCAGUGGAGGGCACCGGGAGCCGCCGUUGUGGCUGCCGUGGUGGCCACUGCCGCCGCGGUGGCGGCAUUUGAGAGCAGAC-UCUGCUGAUGCAGCAGCAGCUGCUGUUGCGGCAAC ((.((.(((.....))).))...))((((((((.((..((((((((((....))))))))))..)).)))...((.(((((.(-(((((((...)))))).))))))).)))))))... ( -66.90, z-score = -2.95, R) >droPer1.super_12 2131043 118 - 2414086 GGACCAGUGGAGGGCACCGGGAGCCGCCGUUGUGGCUGCCGUGGUGGCCACUGCCGCCGCGGUGGCGGCAUUUGAGAGCAGAC-UCUGCUGAUGCAGCAGCAGCUGCUGUUGCGGCAAC ((.((.(((.....))).))...))((((((((.((..((((((((((....))))))))))..)).)))...((.(((((.(-(((((((...)))))).))))))).)))))))... ( -66.90, z-score = -2.95, R) >droWil1.scaffold_180698 5972650 118 - 11422946 CGACCAAUGGAGGGCACUUGGAGCAGCUGUCGUGGCUGCCGUAGUGGCCACUGCCGCUGCAGUAGCAGCAUUCGAAAGUAGAU-UCUGCUGAUGCAGCAAAAGUUGCUGCUGUGGCAGC .........((.(((..........))).))...((((((((((..((.(((...((((((.((((((.((((....)..)))-.)))))).))))))...))).))..)))))))))) ( -51.60, z-score = -1.56, R) >anoGam1.chr2R 34183486 86 + 62725911 -------UGAAUG--AUUUGGGGCUGC-------UUUGGCGUGGGGGCAGUUGGC----------UGCUGUUGGACUGUAACUGUCUUUUGUUGCAAAAUUUGCUGCUGUUG------- -------......--......((((((-------(((......)))))))))(((----------.((.(((....((((((........)))))).)))..)).)))....------- ( -19.90, z-score = 0.43, R) >consensus CGACCAGUGAAUGGAACCCGGAGCCGCC______GUUACCGUUGUGGCCACUGCC____________GCAUUCGAGAGCAGAU_UCUGCUGAUGCAGCAGCAACUGCUGCUGCGGCAAC ...................(..(((((................)))))..).(((.....................(((((....)))))...((((((((....)))))))))))... (-23.41 = -24.41 + 1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:35 2011