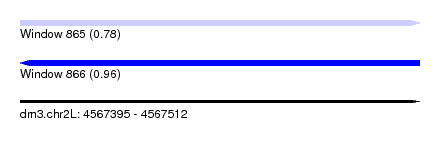
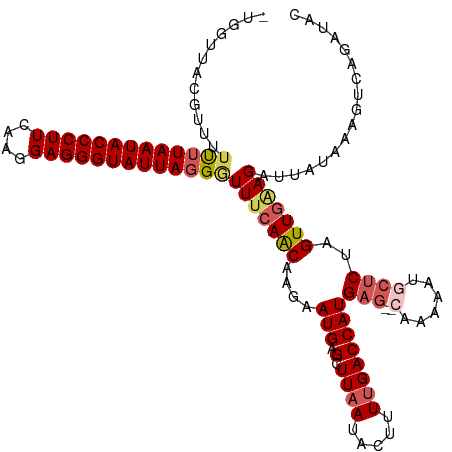
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,567,395 – 4,567,512 |
| Length | 117 |
| Max. P | 0.962640 |

| Location | 4,567,395 – 4,567,512 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.27 |
| Shannon entropy | 0.27435 |
| G+C content | 0.35013 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -14.01 |
| Energy contribution | -14.37 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4567395 117 + 23011544 GUAUCUGACUUUAUAAUCUCUAACUAGGGUACUUUUGUACUCAUGGUCAAAAGUAUUAAGCUCAUUCUUGUUGAAACCCUAAUACCCUCCUUGAAGGGUAUUAAAAAAACUUAAACA- ..........................(((((((((((.((.....)))))))))..........(((.....)))))))(((((((((......)))))))))..............- ( -23.60, z-score = -2.02, R) >droEre2.scaffold_4929 4650868 114 + 26641161 GUAUCUAACUUUAUAAUCUGCAACUAGAGCAUUAUUG--CUCAUGGUCAACAGUAUUAAGCUCAUUCUUGCUGAAAGCAUAAUACCCUCCUUGAAGGGUAUUAAAAAAACGGAACG-- ((.(((............(((.(((((((((....))--))).))))...(((((..((.....))..)))))...)))(((((((((......))))))))).......))))).-- ( -28.60, z-score = -3.50, R) >droYak2.chr2L 4585033 105 + 22324452 --------AUAUCUAA-CUUCAACUAGAGGAUUAUGU--GUCAUGGUCAAGAGUAUAAAGCUCAUUCUUGUUGAAAUCCUAAUACCCUCCUUGAAGGGUAUUAAAAAAACGGAACG-- --------...(((..-.((((((.(((((((((((.--..)))))))..((((.....)))).)))).))))))....(((((((((......))))))))).......)))...-- ( -28.90, z-score = -3.55, R) >droSec1.super_5 2660253 111 + 5866729 ----GUGACUUUAUAAUCUUCAACUAGAGCGUUUUUG--CUCAUGGUCAAAAGUAUUAAGCUCAUUCUUGUUGAAACCCUAAUACCCUCCUUGAAGGGUAUUAAGAAAACGUAACCA- ----(((((((.(((.......(((((((((....))--))).))))......))).))).))))....((((......(((((((((......)))))))))........))))..- ( -23.36, z-score = -1.82, R) >droSim1.chr2L 4453319 116 + 22036055 GUAUCUGACUUUAUAAUCUUCAACUAGAGCGUUUUUG--CUCAUGGUCAAAAGUAUUAAGCUCAUUCUUGUUGAAACCCUAAUACCCUCCUUGAAGGGUAUUAAGAAAACGUAACCAA ...(((............((((((..(((((....))--))).........(((.....))).......))))))....(((((((((......))))))))))))............ ( -23.30, z-score = -1.69, R) >consensus GUAUCUGACUUUAUAAUCUUCAACUAGAGCAUUUUUG__CUCAUGGUCAAAAGUAUUAAGCUCAUUCUUGUUGAAACCCUAAUACCCUCCUUGAAGGGUAUUAAAAAAACGUAACCA_ ..................((((((..((((.............................))))......))))))....(((((((((......)))))))))............... (-14.01 = -14.37 + 0.36)

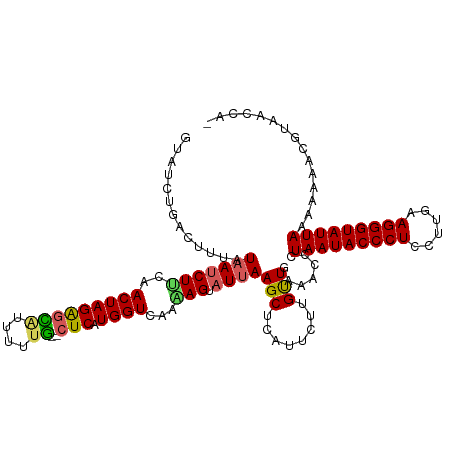
| Location | 4,567,395 – 4,567,512 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.27 |
| Shannon entropy | 0.27435 |
| G+C content | 0.35013 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -19.19 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4567395 117 - 23011544 -UGUUUAAGUUUUUUUAAUACCCUUCAAGGAGGGUAUUAGGGUUUCAACAAGAAUGAGCUUAAUACUUUUGACCAUGAGUACAAAAGUACCCUAGUUAGAGAUUAUAAAGUCAGAUAC -...(((((((((((((((((((((....))))))))))))).(((.....))).))))))))((((((((((.....)).))))))))...........((((....))))...... ( -27.50, z-score = -2.12, R) >droEre2.scaffold_4929 4650868 114 - 26641161 --CGUUCCGUUUUUUUAAUACCCUUCAAGGAGGGUAUUAUGCUUUCAGCAAGAAUGAGCUUAAUACUGUUGACCAUGAG--CAAUAAUGCUCUAGUUGCAGAUUAUAAAGUUAGAUAC --(((((........((((((((((....))))))))))(((.....))).)))))(((((.(((((((.(((...(((--((....)))))..)))))))..))).)))))...... ( -32.30, z-score = -3.40, R) >droYak2.chr2L 4585033 105 - 22324452 --CGUUCCGUUUUUUUAAUACCCUUCAAGGAGGGUAUUAGGAUUUCAACAAGAAUGAGCUUUAUACUCUUGACCAUGAC--ACAUAAUCCUCUAGUUGAAG-UUAGAUAU-------- --............(((((((((((....)))))))))))(((((((((......(((..((((..((........)).--..))))..)))..)))))))-))......-------- ( -24.90, z-score = -2.06, R) >droSec1.super_5 2660253 111 - 5866729 -UGGUUACGUUUUCUUAAUACCCUUCAAGGAGGGUAUUAGGGUUUCAACAAGAAUGAGCUUAAUACUUUUGACCAUGAG--CAAAAACGCUCUAGUUGAAGAUUAUAAAGUCAC---- -...........(((((((((((((....))))))))))))).((((((....(((.(.((((.....))))))))(((--(......))))..))))))((((....))))..---- ( -30.40, z-score = -3.01, R) >droSim1.chr2L 4453319 116 - 22036055 UUGGUUACGUUUUCUUAAUACCCUUCAAGGAGGGUAUUAGGGUUUCAACAAGAAUGAGCUUAAUACUUUUGACCAUGAG--CAAAAACGCUCUAGUUGAAGAUUAUAAAGUCAGAUAC (((..(......(((((((((((((....)))))))))))))(((((((....(((.(.((((.....))))))))(((--(......))))..))))))).......)..))).... ( -30.80, z-score = -2.82, R) >consensus _UGGUUACGUUUUUUUAAUACCCUUCAAGGAGGGUAUUAGGGUUUCAACAAGAAUGAGCUUAAUACUUUUGACCAUGAG__CAAAAAUGCUCUAGUUGAAGAUUAUAAAGUCAGAUAC ............(((((((((((((....)))))))))))))(((((((.(((.((.........(((........)))........)).))).)))))))................. (-19.19 = -19.67 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:43 2011