
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,345,257 – 7,345,397 |
| Length | 140 |
| Max. P | 0.976727 |

| Location | 7,345,257 – 7,345,397 |
|---|---|
| Length | 140 |
| Sequences | 6 |
| Columns | 141 |
| Reading direction | forward |
| Mean pairwise identity | 64.29 |
| Shannon entropy | 0.66691 |
| G+C content | 0.46393 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.08 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.36 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
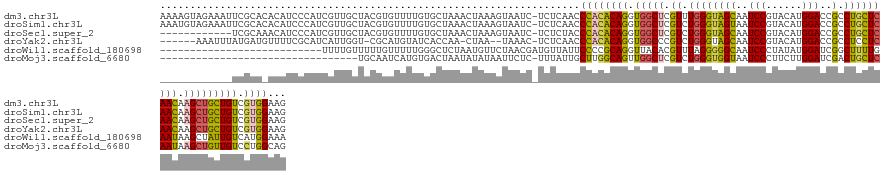
>dm3.chr3L 7345257 140 + 24543557 AAAAGUAGAAAUUCGCACACAUCCCAUCGUUGCUACGUGUUUUGUGCUAAACUAAAGUAAUC-UCUCAACCCACACAGGUGGCUCGUUUGGGUAGCAAUCCGUACAUGGACCGCCUGCUCAACAAGCUGCUGUCGUGGAAG ..............(((((.....((.(((....)))))...)))))...............-.......(((((((.(..(((.(((.(((((((..((((....))))..).))))))))).)))..)))).))))... ( -36.20, z-score = -0.47, R) >droSim1.chr3L 6862708 140 + 22553184 AAAUGUAGAAAUUCGCACACAUCCCAUCGUUGCUACGUGUUUUGUGCUAAACUAAAGUAAUC-UCUCAACCCACACAGGUGGCUCGUCUGGGUAGUAAUCCGUACAUGGACCGCCUGCUCAACAAGCUGCUGUCGUGGAAG ...((.(((.....(((((.....((.(((....)))))...)))))...((....))....-)))))..(((((((.(..(((.((.(((((((...((((....))))....))))))))).)))..)))).))))... ( -35.70, z-score = -0.46, R) >droSec1.super_2 7269938 128 + 7591821 ------------UCGCAAACAUCCCAUCGUUGCUACGUGUUUUGUGCUAAACUAAAGUAAUC-UCUCUACCCACACAGGUGGCUCGUCUGGGUAGCAAUCCGUACAUGGACCGCCUGCUCAACAAGCUGCUGUCGUGGAAG ------------................((((((....((((......))))...)))))).-.......(((((((.(..(((.((.((((((((..((((....))))..).))))))))).)))..)))).))))... ( -33.10, z-score = -0.32, R) >droYak2.chr3L 7950132 130 + 24197627 ------AAAUUUAUGAUGUUUUCGCAUCAUUGGU-CGCAUGUAUCACCAA-CUAA--UAAAC-UCUCAACCCACACAGGUGGCCCGUCUGGGUAGCAAUCCGUACAUGGACCGCCUCCUCAACAAGCUGCUGUCGUGGAAG ------......(((((((....))))))).(((-(.(((((((((((..-....--.....-..............))))((((....))))........)))))))))))...(((.(.(((......))).).))).. ( -33.11, z-score = -0.81, R) >droWil1.scaffold_180698 9209510 114 + 11422946 ---------------------------UUUUGUUUUUGUUUUUGGGCUCUAAUGUUCUAACGAUGUUAUUCCCCGCAGGUUACACGUUUAGGGGGCAAUCCCUAUAUGGAUCGGCUUUUGAAUAAGCUAUUGUCAUGGAAA ---------------------------........(((((...((((......)))).))))).....((((..((((......(((.((((((....)))))).)))....(((((......))))).))))...)))). ( -24.90, z-score = 0.30, R) >droMoj3.scaffold_6680 5682094 107 + 24764193 ---------------------------------UGCAAUCAUGUGACUAAUAUAUAAUUCUC-UUUAUUGCUUGGCAGUUGGCUCGUCUGGGUGGUAAUCCCUUCUUGGAUCGACUGCUCAAUAAGCUGUUGUCCUGGCAG ---------------------------------(((....((((.....))))(((((....-(((((((...(((((((((..((...(((.......)))....))..))))))))))))))))..)))))....))). ( -25.30, z-score = -0.38, R) >consensus ____________UCGCA_ACAUCCCAUCGUUGCUACGUGUUUUGUGCUAAACUAAAGUAAUC_UCUCAACCCACACAGGUGGCUCGUCUGGGUAGCAAUCCGUACAUGGACCGCCUGCUCAACAAGCUGCUGUCGUGGAAG ......................................................................((((((((.(((((.((.((((((((..(((......)))..).))))))))).))))))))).))))... (-20.55 = -20.08 + -0.47)

| Location | 7,345,257 – 7,345,397 |
|---|---|
| Length | 140 |
| Sequences | 6 |
| Columns | 141 |
| Reading direction | reverse |
| Mean pairwise identity | 64.29 |
| Shannon entropy | 0.66691 |
| G+C content | 0.46393 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -24.19 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
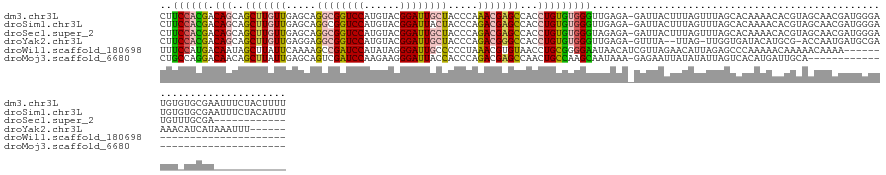
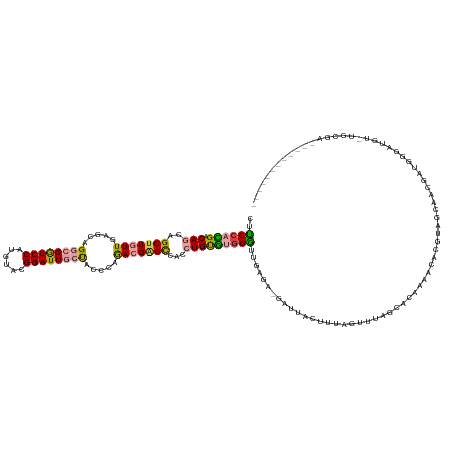
>dm3.chr3L 7345257 140 - 24543557 CUUCCACGACAGCAGCUUGUUGAGCAGGCGGUCCAUGUACGGAUUGCUACCCAAACGAGCCACCUGUGUGGGUUGAGA-GAUUACUUUAGUUUAGCACAAAACACGUAGCAACGAUGGGAUGUGUGCGAAUUUCUACUUUU ..(((((.((((..(((((((.....((((((((......)))))))).....)))))))...)))))))))...(((-((((((....))...(((((...(((((....))).)).....))))).)))))))...... ( -43.90, z-score = -1.65, R) >droSim1.chr3L 6862708 140 - 22553184 CUUCCACGACAGCAGCUUGUUGAGCAGGCGGUCCAUGUACGGAUUACUACCCAGACGAGCCACCUGUGUGGGUUGAGA-GAUUACUUUAGUUUAGCACAAAACACGUAGCAACGAUGGGAUGUGUGCGAAUUUCUACAUUU ..(((((.((((..(((((((.....((..((((......))))..)).....)))))))...))))))))).(((((-((((((....))...(((((...(((((....))).)).....))))).))))))).))... ( -40.30, z-score = -0.99, R) >droSec1.super_2 7269938 128 - 7591821 CUUCCACGACAGCAGCUUGUUGAGCAGGCGGUCCAUGUACGGAUUGCUACCCAGACGAGCCACCUGUGUGGGUAGAGA-GAUUACUUUAGUUUAGCACAAAACACGUAGCAACGAUGGGAUGUUUGCGA------------ ......((((((....)))))).(((((((..(((((((((((((.(((((((.(((.......))).)))))))...-))))......((((......)))).))).)))....)))..)))))))..------------ ( -37.70, z-score = -0.55, R) >droYak2.chr3L 7950132 130 - 24197627 CUUCCACGACAGCAGCUUGUUGAGGAGGCGGUCCAUGUACGGAUUGCUACCCAGACGGGCCACCUGUGUGGGUUGAGA-GUUUA--UUAG-UUGGUGAUACAUGCG-ACCAAUGAUGCGAAAACAUCAUAAAUUU------ ..(((((.((((..(((((((..((.((((((((......)))))))).))..)))))))...)))))))))....((-(((((--(..(-(((((..........-))))))((((......))))))))))))------ ( -44.80, z-score = -2.65, R) >droWil1.scaffold_180698 9209510 114 - 11422946 UUUCCAUGACAAUAGCUUAUUCAAAAGCCGAUCCAUAUAGGGAUUGCCCCCUAAACGUGUAACCUGCGGGGAAUAACAUCGUUAGAACAUUAGAGCCCAAAAACAAAAACAAAA--------------------------- ....((((......((((......)))).........(((((......)))))..)))).........(((..(((..((....))...)))...)))................--------------------------- ( -17.00, z-score = 0.48, R) >droMoj3.scaffold_6680 5682094 107 - 24764193 CUGCCAGGACAACAGCUUAUUGAGCAGUCGAUCCAAGAAGGGAUUACCACCCAGACGAGCCAACUGCCAAGCAAUAAA-GAGAAUUAUAUAUUAGUCACAUGAUUGCA--------------------------------- ......((......(((..((.....(..(((((......)))))..).....))..)))......))..(((((...-(.((.(((.....))))).)...))))).--------------------------------- ( -16.60, z-score = 0.48, R) >consensus CUUCCACGACAGCAGCUUGUUGAGCAGGCGGUCCAUGUACGGAUUGCUACCCAGACGAGCCACCUGUGUGGGUUGAGA_GAUUACUUUAGUUUAGCACAAAACACGUAGCAACGAUGGGAUGU_UGCGA____________ ..((((((.(((..(((((((.....((((((((......)))))))).....)))))))...)))))))))..................................................................... (-24.19 = -24.28 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:27 2011