| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,567,093 – 4,567,183 |
| Length | 90 |
| Max. P | 0.621465 |

| Location | 4,567,093 – 4,567,183 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Shannon entropy | 0.34339 |
| G+C content | 0.49474 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
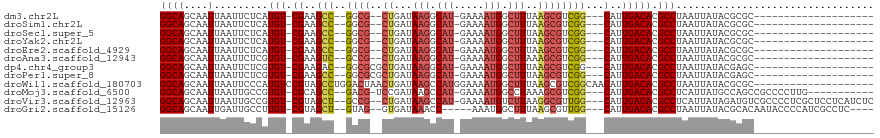
>dm3.chr2L 4567093 90 - 23011544 GGCAGCAAUUAAUUCUCAUGU-CGAAGCC--GGCG--CUGAUAAGGCAU-GAAAAUGGCUUUAAGCGUCGG---CAUUGACACGCCUAAUUAUACGCGC-------------------- .((.((...(((((..(.(((-(((.(((--((((--((...(((.(((-....))).)))..))))))))---).)))))).)...)))))...))))-------------------- ( -36.50, z-score = -4.15, R) >droSim1.chr2L 4453020 90 - 22036055 GGCAGCAAUUAAUUCUCAUGU-CGAAGCC--GGCG--CUGAUAAGGCAU-GAAAAUGGCUUUAAGCGUCGG---CAUUGACACGCCUAAUUAUACGCGC-------------------- .((.((...(((((..(.(((-(((.(((--((((--((...(((.(((-....))).)))..))))))))---).)))))).)...)))))...))))-------------------- ( -36.50, z-score = -4.15, R) >droSec1.super_5 2659946 90 - 5866729 GGCAGCAAUUAAUUCUCAUGU-CGAAGCC--GGCG--CUGAUAAGGCAU-GAAAAUGGCUUUAAGCGUCGG---CAUUGACACGCCUAAUUAUACGCGC-------------------- .((.((...(((((..(.(((-(((.(((--((((--((...(((.(((-....))).)))..))))))))---).)))))).)...)))))...))))-------------------- ( -36.50, z-score = -4.15, R) >droYak2.chr2L 4584737 90 - 22324452 GGCAGCAAUUAAUUCUCAUGU-CGAAGCC--GGCG--CUGAUAAGGCAU-GAAAAUGGCUUUAAGCGUCGG---CAUUGACACGCCUAAUUAUACGCGC-------------------- .((.((...(((((..(.(((-(((.(((--((((--((...(((.(((-....))).)))..))))))))---).)))))).)...)))))...))))-------------------- ( -36.50, z-score = -4.15, R) >droEre2.scaffold_4929 4650576 90 - 26641161 GGCAGCAAUUAAUUCUCAUGU-CGAAGCC--GGCG--CUGAUAAGGCAU-GAAAAUGGCUUUAAGCGUCGG---CAUUGACACGCCUAAUUAUACGCGC-------------------- .((.((...(((((..(.(((-(((.(((--((((--((...(((.(((-....))).)))..))))))))---).)))))).)...)))))...))))-------------------- ( -36.50, z-score = -4.15, R) >droAna3.scaffold_12943 1766058 90 + 5039921 GGCAGCAAUUAAUUCUCGUGU-CGAAGUC--GCCG--CUGAUAAGGCAU-GAAAAUGGCUUAAAGCGUCGG---CAUUGACACGCCUAAUUAUACGCGC-------------------- .((.((...(((((..(((((-(((.(((--(.((--((..((((.(((-....))).)))).)))).)))---).))))))))...)))))...))))-------------------- ( -35.80, z-score = -3.91, R) >dp4.chr4_group3 404816 92 + 11692001 GGCAGCAAUUAAUUCUCGUGU-CGAAGAC--GGCGCGCUGAUAAGGCAU-GAAAAUGGCUUUAAGCGUCGG---CAUUGACACGCCUAAUUAUACGAGC-------------------- ((((....)........((((-(((.(.(--(((((......(((.(((-....))).)))...)))))).---).)))))))))).............-------------------- ( -29.30, z-score = -1.64, R) >droPer1.super_8 1451469 92 + 3966273 GGCAGCAAUUAAUUCUCGUGU-CGAAGCC--GGCGCGCUGAUAAGGCAU-GAAAAUGGCUUUAAGCGUCGG---CAUUGACACGCCUAAUUAUACGAGC-------------------- ((((....)........((((-(((.(((--(((((......(((.(((-....))).)))...)))))))---).)))))))))).............-------------------- ( -36.40, z-score = -3.45, R) >droWil1.scaffold_180703 1309005 99 + 3946847 GGCAGCAAUUAAUUCCCAUGUCCGUAGCCUGGACUAACUGAUAAGCCAUGGAAAAUGGCUUUAAGCGUCGGCAACAUUGACACGCCUAAUUAUACGCGC-------------------- (((.((.............(((((.....)))))........(((((((.....)))))))...))(((((.....)))))..))).............-------------------- ( -24.60, z-score = -0.48, R) >droMoj3.scaffold_6500 11930042 100 + 32352404 GGCAGCAAUUAAUUGCCGUGU-CGCAGCC--GACG-UCCGAUAAGCCAU-GAAAAUGGCCUAAAGCGUCGG---CAUUGACACGCCUCAUUAUGCCAGCCGCCCCUUG----------- ((((((((....))))(((((-((..(((--((((-(....((.(((((-....))))).))..)))))))---)..)))))))........))))............----------- ( -41.30, z-score = -4.84, R) >droVir3.scaffold_12963 8478251 110 + 20206255 GGCAGCAAUUAAUUGCCGUGU-CGUAGCU--GCCG--CUGAUAAGCCAU-GAAAAUGUCUUAAGGCGUUGG---CAUUGACACGCCUCAUUAUAGAUGUCGCCCCUCGCUCCUCAUCUC (((.((((....)))).((((-((..(((--..((--((..((((.(((-....))).)))).))))..))---)..))))))))).......(((((..((.....))....))))). ( -36.60, z-score = -2.90, R) >droGri2.scaffold_15126 3119001 102 + 8399593 GGCAGCAAUUGAUUGCCUUGU-CGUAGCU--GUAG--GUGAUAAACC-----AAAUGGCUUUAAGCGUUGG---CAUUGACACGCCUAAUUAUACGCACAAUACCCCAUCGCCUC---- (((.((((....)))).((((-((((...--.(((--(((.((((((-----....)).)))).((....)---).......))))))....)))).)))).........)))..---- ( -26.30, z-score = -0.62, R) >consensus GGCAGCAAUUAAUUCUCAUGU_CGAAGCC__GGCG__CUGAUAAGGCAU_GAAAAUGGCUUUAAGCGUCGG___CAUUGACACGCCUAAUUAUACGCGC____________________ (((................((....((((.........((......))........))))....))(((((.....)))))..)))................................. ( -9.62 = -9.70 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:41 2011