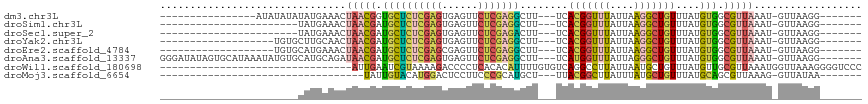
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,247,564 – 7,247,654 |
| Length | 90 |
| Max. P | 0.956139 |

| Location | 7,247,564 – 7,247,654 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.30 |
| Shannon entropy | 0.53373 |
| G+C content | 0.40921 |
| Mean single sequence MFE | -16.37 |
| Consensus MFE | -7.45 |
| Energy contribution | -7.18 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.88 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
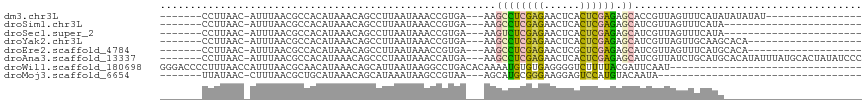
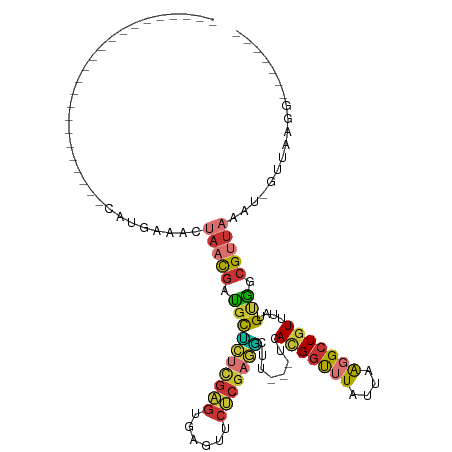
>dm3.chr3L 7247564 90 + 24543557 -------CCUUAAC-AUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGA---AAGCCUCGAGAACUCACUCGAGAGCACCGUUAGUUUCAUAUAUAUAU---------------- -------.......-..((((((.(((....................))).---..((((((((......)))))).))..))))))..............---------------- ( -15.65, z-score = -2.97, R) >droSim1.chr3L 6759520 83 + 22553184 -------CCUUAAC-AUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGA---AAGCCUCGAGAACUCACUCGAGAGCAUCGUUAGUUUCAUA----------------------- -------.......-..((((((.(((....................))).---..((((((((......)))))).))..)))))).......----------------------- ( -15.45, z-score = -2.61, R) >droSec1.super_2 7173822 83 + 7591821 -------CCUUAAC-AUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGA---AAGUCUCGAGAACUCACUCGAGAGCAUCGUUAGUUUCAUA----------------------- -------.......-..((((((.(((....................))).---...(((((((......)))))))....)))))).......----------------------- ( -13.55, z-score = -1.61, R) >droYak2.chr3L 7849026 87 + 24197627 -------CCUUAAC-AUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGA---AAGCCUCGAGAACUCACUCGAGAGCAUCGUUAGUUGCAAGCACA------------------- -------......(-(.((((((.(((....................))).---..((((((((......)))))).))..)))))).))........------------------- ( -16.45, z-score = -2.00, R) >droEre2.scaffold_4784 21666111 87 - 25762168 -------CCUUAAC-AUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGA---AAGCCUCGAGAACUCGCUCGAGAGCAUCGUUAGUUUCAUGCACA------------------- -------.......-..((((((.(((....................))).---..((((((((......)))))).))..))))))...........------------------- ( -15.65, z-score = -1.45, R) >droAna3.scaffold_13337 14704462 106 + 23293914 -------CCUUAAC-AUUUAACGCCACAUAAACAGCCCUAAUAAACCAUGA---AAGCCUCGAGAACUCACUCGAGAGCAUCGUUAUCUGCAUGCACAUAUUUAUGCACUAUAUCCC -------.......-...((((((..........))...............---..((((((((......)))))).))...))))..((((((........))))))......... ( -17.40, z-score = -2.48, R) >droWil1.scaffold_180698 9080171 85 + 11422946 GGGACCCCUUUAACCAUUUAACGCAACAUAAACAGCAUUAAUAAGGCCUGACACAAAAUGUGUGAGGGGUCUUUUACGAUUCAAU-------------------------------- ((((((((((...((..((((.((..........)).))))...))....((((.....))))))))))))))............-------------------------------- ( -20.90, z-score = -2.14, R) >droMoj3.scaffold_6654 1065197 72 - 2564135 -------UUAUAAC-CUUUAACGCUGCAUAAACAGCAUAAAUAAGCCGUAA---AGCAUGCGGGAAGGAGUCCAUGUACAAUA---------------------------------- -------......(-((((...((((......)))).........(((((.---....))))))))))...............---------------------------------- ( -15.90, z-score = -1.45, R) >consensus _______CCUUAAC_AUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGA___AAGCCUCGAGAACUCACUCGAGAGCAUCGUUAGUUUCAUA_______________________ ........................................................((((((((......)))))).))...................................... ( -7.45 = -7.18 + -0.28)
| Location | 7,247,564 – 7,247,654 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.30 |
| Shannon entropy | 0.53373 |
| G+C content | 0.40921 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -12.23 |
| Energy contribution | -12.45 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
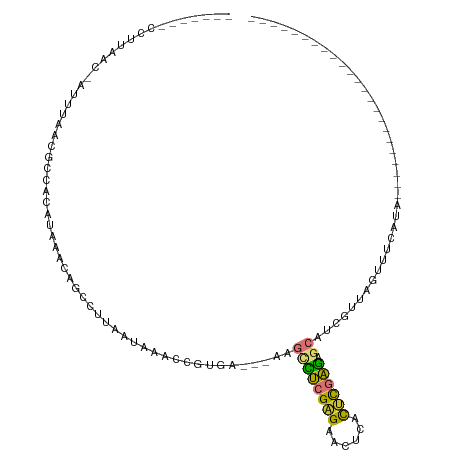
>dm3.chr3L 7247564 90 - 24543557 ----------------AUAUAUAUAUGAAACUAACGGUGCUCUCGAGUGAGUUCUCGAGGCUU---UCACGGUUUAUUAAGGCUGUUUAUGUGGCGUUAAAU-GUUAAGG------- ----------------...........(((((......((.((((((......))))))))..---....))))).((((.((..(....)..)).))))..-.......------- ( -20.80, z-score = -1.01, R) >droSim1.chr3L 6759520 83 - 22553184 -----------------------UAUGAAACUAACGAUGCUCUCGAGUGAGUUCUCGAGGCUU---UCACGGUUUAUUAAGGCUGUUUAUGUGGCGUUAAAU-GUUAAGG------- -----------------------........((((((.((.((((((......))))))))..---))........((((.((..(....)..)).))))..-))))...------- ( -21.10, z-score = -1.43, R) >droSec1.super_2 7173822 83 - 7591821 -----------------------UAUGAAACUAACGAUGCUCUCGAGUGAGUUCUCGAGACUU---UCACGGUUUAUUAAGGCUGUUUAUGUGGCGUUAAAU-GUUAAGG------- -----------------------........((((((...(((((((......)))))))...---))........((((.((..(....)..)).))))..-))))...------- ( -19.90, z-score = -1.12, R) >droYak2.chr3L 7849026 87 - 24197627 -------------------UGUGCUUGCAACUAACGAUGCUCUCGAGUGAGUUCUCGAGGCUU---UCACGGUUUAUUAAGGCUGUUUAUGUGGCGUUAAAU-GUUAAGG------- -------------------....((((((..(((((..((.((((((......))))))))..---..(((((((....)))))))........)))))..)-).)))).------- ( -22.90, z-score = -0.99, R) >droEre2.scaffold_4784 21666111 87 + 25762168 -------------------UGUGCAUGAAACUAACGAUGCUCUCGAGCGAGUUCUCGAGGCUU---UCACGGUUUAUUAAGGCUGUUUAUGUGGCGUUAAAU-GUUAAGG------- -------------------...((((.(((((...((.((.((((((......))))))))..---))..))))).((((.((..(....)..)).))))))-)).....------- ( -21.30, z-score = -0.37, R) >droAna3.scaffold_13337 14704462 106 - 23293914 GGGAUAUAGUGCAUAAAUAUGUGCAUGCAGAUAACGAUGCUCUCGAGUGAGUUCUCGAGGCUU---UCAUGGUUUAUUAGGGCUGUUUAUGUGGCGUUAAAU-GUUAAGG------- ........(((((((....))))))).....((((((.((.((((((......))))))))..---))........((((.((..(....)..)).))))..-))))...------- ( -27.60, z-score = -0.99, R) >droWil1.scaffold_180698 9080171 85 - 11422946 --------------------------------AUUGAAUCGUAAAAGACCCCUCACACAUUUUGUGUCAGGCCUUAUUAAUGCUGUUUAUGUUGCGUUAAAUGGUUAAAGGGGUCCC --------------------------------..............(((((((.((((.....))))..((((...(((((((..........)))))))..))))..))))))).. ( -24.80, z-score = -2.74, R) >droMoj3.scaffold_6654 1065197 72 + 2564135 ----------------------------------UAUUGUACAUGGACUCCUUCCCGCAUGCU---UUACGGCUUAUUUAUGCUGUUUAUGCAGCGUUAAAG-GUUAUAA------- ----------------------------------...............((((..(((.(((.---..(((((........)))))....))))))...)))-)......------- ( -17.10, z-score = -1.28, R) >consensus _______________________CAUGAAACUAACGAUGCUCUCGAGUGAGUUCUCGAGGCUU___UCACGGUUUAUUAAGGCUGUUUAUGUGGCGUUAAAU_GUUAAGG_______ ...............................(((((.((((((((((......)))))))........(((((((....)))))))....))).))))).................. (-12.23 = -12.45 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:20 2011