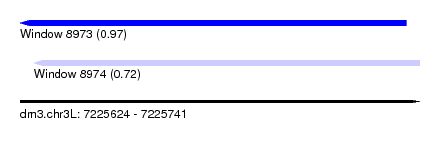
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,225,624 – 7,225,741 |
| Length | 117 |
| Max. P | 0.969332 |

| Location | 7,225,624 – 7,225,737 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.19 |
| Shannon entropy | 0.13095 |
| G+C content | 0.39072 |
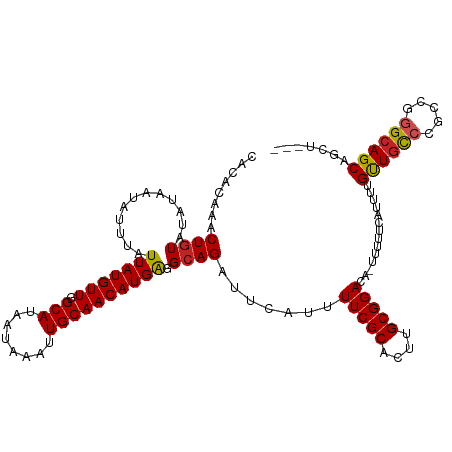
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -29.63 |
| Energy contribution | -29.46 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
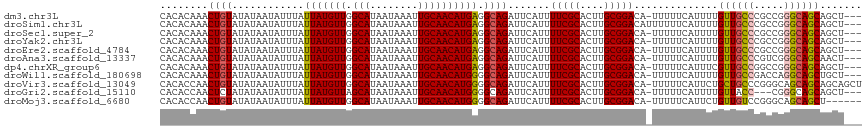
>dm3.chr3L 7225624 113 - 24543557 CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCUGCUU--- ....................((((((((.((........))))))))))(((((.......(((((....)))))..-.........(((((((((...)))))))))))))).--- ( -32.20, z-score = -2.18, R) >droSim1.chr3L 6737425 114 - 22553184 CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACAUUUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCUGCUU--- ....................((((((((.((........))))))))))(((((.......(((((....)))))............(((((((((...)))))))))))))).--- ( -32.20, z-score = -2.20, R) >droSec1.super_2 7151900 113 - 7591821 CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCUGCUU--- ....................((((((((.((........))))))))))(((((.......(((((....)))))..-.........(((((((((...)))))))))))))).--- ( -32.20, z-score = -2.18, R) >droYak2.chr3L 7825930 113 - 24197627 CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCUGCUU--- ....................((((((((.((........))))))))))(((((.......(((((....)))))..-.........(((((((((...)))))))))))))).--- ( -32.20, z-score = -2.18, R) >droEre2.scaffold_4784 21644254 113 + 25762168 CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCUGCUU--- ....................((((((((.((........))))))))))(((((.......(((((....)))))..-.........(((((((((...)))))))))))))).--- ( -32.20, z-score = -2.18, R) >droAna3.scaffold_13337 1750259 113 - 23293914 CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGUCGGGCAGCAACUGCUU--- ....................((((((((.((........))))))))))(((((.......(((((....)))))..-.........(((((((((...)))))))))))))).--- ( -32.80, z-score = -3.20, R) >dp4.chrXR_group6 3987125 113 - 13314419 CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUCGUUGCCGGCCGGGCAGCAGCUGCUC--- ....................((((((((.((........))))))))))(((((.......(((((....)))))..-...........((((((.....))))))..))))).--- ( -29.70, z-score = -1.51, R) >droWil1.scaffold_180698 7828918 113 - 11422946 CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGGGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCGACCAGGCAGCUGCUGCUA--- ....((((.......((((((((((....))))))))))......))))(((((.......(((((....)))))..-...........((((((.....))))))..))))).--- ( -30.02, z-score = -1.74, R) >droVir3.scaffold_13049 9468106 116 + 25233164 CCAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGGGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUCUGCUGCCCGGGCAGCAGCAGCAGCUGCUU (((..((((......((((((((((....))))))))))))))...)))(((((.......(((((....)))))..-.........(((((((....)))))))......))))). ( -34.90, z-score = -1.88, R) >droGri2.scaffold_15110 12873453 110 - 24565398 CCAACUCUAUAUAAUAUUUAUUAUGUUAGCAUAAUAAAUUGCAACAUGGGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUACC---CGGGCAGCAGCUGCUU--- ((..((((((.....((((((((((....))))))))))......))))))..((...((.(((((....))))).)-)...))...........---.))((((...))))..--- ( -23.10, z-score = -0.85, R) >droMoj3.scaffold_6680 21058008 110 - 24764193 CCAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGGGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUCUGUUGUC---CGGGCAGCAGCUGCUC--- (((..((((......((((((((((....))))))))))))))...)))(((((.......(((((....)))))..-.........(((((((.---...)))))))))))).--- ( -30.40, z-score = -2.26, R) >consensus CAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA_UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCUGCUU___ ....................(((((((.(((........))))))))))(((((.......(((((....)))))............((((((((.....))))))))))))).... (-29.63 = -29.46 + -0.17)
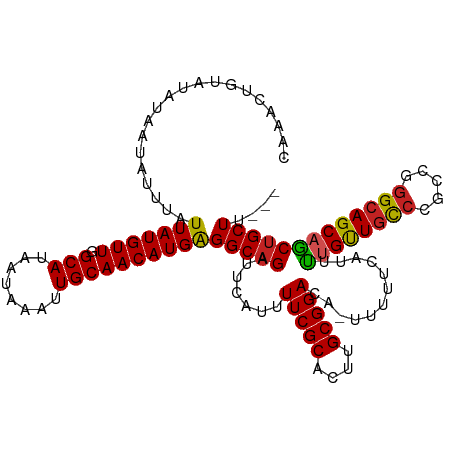
| Location | 7,225,628 – 7,225,741 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.99 |
| Shannon entropy | 0.13369 |
| G+C content | 0.38909 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -24.44 |
| Energy contribution | -24.41 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7225628 113 - 24543557 CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCU--- ........................((((((((.((........))))))))))(((.((...((.(((((....))))).)-)...))....((((((((...)))))))))))--- ( -29.00, z-score = -1.80, R) >droSim1.chr3L 6737429 114 - 22553184 CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACAUUUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCU--- ........((((............((((((((.((........)))))))))).)))).......(((((....)))))............(((((((((...)))))))))..--- ( -28.62, z-score = -1.68, R) >droSec1.super_2 7151904 113 - 7591821 CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCU--- ........................((((((((.((........))))))))))(((.((...((.(((((....))))).)-)...))....((((((((...)))))))))))--- ( -29.00, z-score = -1.80, R) >droYak2.chr3L 7825934 113 - 24197627 CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCU--- ........................((((((((.((........))))))))))(((.((...((.(((((....))))).)-)...))....((((((((...)))))))))))--- ( -29.00, z-score = -1.80, R) >droEre2.scaffold_4784 21644258 113 + 25762168 CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCU--- ........................((((((((.((........))))))))))(((.((...((.(((((....))))).)-)...))....((((((((...)))))))))))--- ( -29.00, z-score = -1.80, R) >droAna3.scaffold_13337 1750263 113 - 23293914 CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCCGUCGGGCAGCAACU--- ........((((............((((((((.((........)))))))))).)))).......(((((....)))))..-.........(((((((((...)))))))))..--- ( -28.92, z-score = -2.49, R) >dp4.chrXR_group6 3987129 113 - 13314419 CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUCGUUGCCGGCCGGGCAGCAGCU--- ........................((((((((.((........))))))))))(((.((...((.(((((....))))).)-)...)).....((((((.....)))))).)))--- ( -26.80, z-score = -1.17, R) >droWil1.scaffold_180698 7828922 113 - 11422946 CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGGGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUGCCGACCAGGCAGCUGCU--- ........((((.......((((((((((....))))))))))......))))((((((...((.(((((....))))).)-)...)).....((((((.....))))))))))--- ( -27.12, z-score = -1.40, R) >droVir3.scaffold_13049 9468110 116 + 25233164 CACACCAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGGGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUCUGCUGCCCGGGCAGCAGCAGCAGCU ....(((..((((......((((((((((....))))))))))))))...)))(((.((...((.(((((....))))).)-)...))...(((((((....))))))).....))) ( -32.50, z-score = -1.83, R) >droGri2.scaffold_15110 12873457 110 - 24565398 CACACCAACUCUAUAUAAUAUUUAUUAUGUUAGCAUAAUAAAUUGCAACAUGGGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUUUGUUACC---CGGGCAGCAGCU--- ...................((((((((((....))))))))))(((....(((((((((......(((((....)))))..-........)))))..))---)).)))......--- ( -21.59, z-score = -1.17, R) >droMoj3.scaffold_6680 21058012 110 - 24764193 CACACCAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGGGGCAGAUUCAUUUUCGCACUUGCGGACA-UUUUUCAUUCUGUUGUCCGGGCAGCAGCU------ ....(((..((((......((((((((((....))))))))))))))...)))(((.((...((.(((((....))))).)-)...))....((((((....)))))))))------ ( -25.80, z-score = -1.25, R) >consensus CACACAAACUGUAUAUAAUAUUUAUUAUGUUGGCAUAAUAAAUUGCAACAUGAGGCAGAUUCAUUUUCGCACUUGCGGACA_UUUUUCAUUUUGUUGCCCGCCGGGCAGCAGCU___ ........((((............(((((((.(((........)))))))))).)))).......(((((....)))))..............((((((.....))))))....... (-24.44 = -24.41 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:18 2011