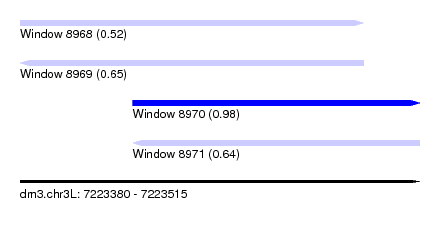
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,223,380 – 7,223,515 |
| Length | 135 |
| Max. P | 0.979652 |

| Location | 7,223,380 – 7,223,496 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.01 |
| Shannon entropy | 0.59078 |
| G+C content | 0.55289 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -15.97 |
| Energy contribution | -18.18 |
| Covariance contribution | 2.21 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7223380 116 + 24543557 GACGAAGGGGUGAGGGG--GGUGGACAUUUUAACAACCCAGGAUCGCAGGAUACUAGAAUCCCAGCAUUGCAACAUCCCAGCUUUCCAACAUCCUGGCGGCACAAGUUGCCAGUUGCC ......((((((.(((.--.((..........))..)))......((.((((......))))..)).......))))))..............((((((((....))))))))..... ( -35.00, z-score = -0.12, R) >droSim1.chr3L 6734907 114 + 22553184 GACGAAGGGGAGGGG-GUUGGGGGGCAUUUUAAC---CCAGGAUCGCAGGAUCCUAGAAUCCCAGCAUUGCAACAUCCCAGCUUUCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCC ......((..(((((-(((((((((........)---))((((((....)))))).....))))((...))...)))))..))..)).(((..((((((((....)))))))).))). ( -45.20, z-score = -1.61, R) >droSec1.super_2 7149617 118 + 7591821 GACGAAGGGGUGGGGAGUGGGGGGGCAUUUUAACAACCCAGGAUCGCAGGAUCCUAGAAUCCCAGCAGUGCAACAUCCCAGCUUUCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCC ......((((((((((..(((..(.........)..)))((((((....))))))....)))).((...))..)))))).........(((..((((((((....)))))))).))). ( -45.90, z-score = -1.34, R) >droYak2.chr3L 7823872 108 + 24197627 --------GAUGAAGGG--GAUGGGCAUUUUAACAACCCAGGAUCGCUGGAUCCUGGAAUCCCAACAACCCAACAUCCCAGCUUCACAGCAUCCUGGCGGCACAAGUUGCCAGUUGCC --------..(((((((--((((((............((((((((....))))))))...........))))...))))..)))))..(((..((((((((....)))))))).))). ( -42.50, z-score = -3.18, R) >droEre2.scaffold_4784 21642326 100 - 25762168 --------GAUGAAGGG--GGUGGGCAUUUUAACAACCCAGGAUCGCAGCAUCCUGGAAUCCCAACA--------UCCCAGCUUCCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCC --------((((..(((--((((((............(((((((......)))))))..........--------.)))).)))))...))))((((((((....))))))))..... ( -36.90, z-score = -1.51, R) >droWil1.scaffold_180698 7826472 87 + 11422946 -------UACACAGGGGUGGGGGGGCUGUCUAUUAAAGUGGG--UGUGGGUAACUGGGGUAAUAACAG-----------AACUUGCCAAUGU--UGCUAGUGUAAGGCA--------- -------...(((..((..((....((((.(((((...(.((--(.......))).)..)))))))))-----------..))..))..)))--((((.......))))--------- ( -20.50, z-score = 0.36, R) >consensus _______GGGUGAGGGG__GGGGGGCAUUUUAACAACCCAGGAUCGCAGGAUCCUAGAAUCCCAACAGUGCAACAUCCCAGCUUUCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCC ...................((..(((.............((((((....)))))).(........)..............)))..)).(((..((((((((....)))))))).))). (-15.97 = -18.18 + 2.21)

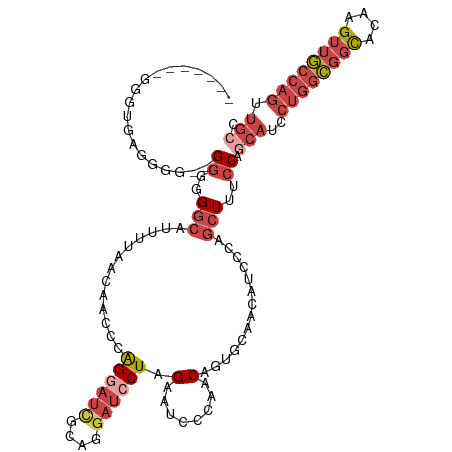
| Location | 7,223,380 – 7,223,496 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.01 |
| Shannon entropy | 0.59078 |
| G+C content | 0.55289 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -21.35 |
| Energy contribution | -23.92 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.48 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7223380 116 - 24543557 GGCAACUGGCAACUUGUGCCGCCAGGAUGUUGGAAAGCUGGGAUGUUGCAAUGCUGGGAUUCUAGUAUCCUGCGAUCCUGGGUUGUUAAAAUGUCCACC--CCCCUCACCCCUUCGUC ((((((.((((.....)))).(((((((((.(((..((((((((.((((...)).)).)))))))).))).)).)))))))))))))............--................. ( -35.30, z-score = -0.03, R) >droSim1.chr3L 6734907 114 - 22553184 GGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGAAAGCUGGGAUGUUGCAAUGCUGGGAUUCUAGGAUCCUGCGAUCCUGG---GUUAAAAUGCCCCCCAAC-CCCCUCCCCUUCGUC ((((...(....)...))))....(((.(((....))).(((..((......))((((.....((((((....))))))((---((......))))))))..-.))))))........ ( -37.10, z-score = -0.07, R) >droSec1.super_2 7149617 118 - 7591821 GGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGAAAGCUGGGAUGUUGCACUGCUGGGAUUCUAGGAUCCUGCGAUCCUGGGUUGUUAAAAUGCCCCCCCACUCCCCACCCCUUCGUC ((((...(....)...))))...(((..(((....)))((((..((......))((((.....((((((....))))))((((.........)))).))))...))))..)))..... ( -36.60, z-score = 0.57, R) >droYak2.chr3L 7823872 108 - 24197627 GGCAACUGGCAACUUGUGCCGCCAGGAUGCUGUGAAGCUGGGAUGUUGGGUUGUUGGGAUUCCAGGAUCCAGCGAUCCUGGGUUGUUAAAAUGCCCAUC--CCCUUCAUC-------- ((((.(((((..........)))))..))))((((((..((((...(((((..((((.(..((((((((....))))))))..).))))...)))))))--)))))))).-------- ( -46.60, z-score = -2.79, R) >droEre2.scaffold_4784 21642326 100 + 25762168 GGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGGAAGCUGGGA--------UGUUGGGAUUCCAGGAUGCUGCGAUCCUGGGUUGUUAAAAUGCCCACC--CCCUUCAUC-------- ((((...(....)...)))).((((....))))((((..(((.--------....((((((.(((....))).))))))((((.........)))).))--).))))...-------- ( -36.90, z-score = -0.59, R) >droWil1.scaffold_180698 7826472 87 - 11422946 ---------UGCCUUACACUAGCA--ACAUUGGCAAGUU-----------CUGUUAUUACCCCAGUUACCCACA--CCCACUUUAAUAGACAGCCCCCCCACCCCUGUGUA------- ---------.....(((((.....--.....(((....(-----------((((((........((.....)).--.......)))))))..)))...........)))))------- ( -10.19, z-score = 0.00, R) >consensus GGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGAAAGCUGGGAUGUUGCAAUGCUGGGAUUCCAGGAUCCUGCGAUCCUGGGUUGUUAAAAUGCCCACC__CCCCUCACCC_______ ((((...((((.....)))).(((((((((.(((...(((((((.((........)).)))))))..))).)).)))))))..........))))....................... (-21.35 = -23.92 + 2.56)

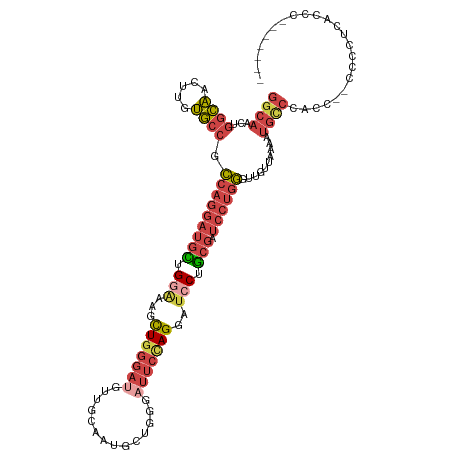
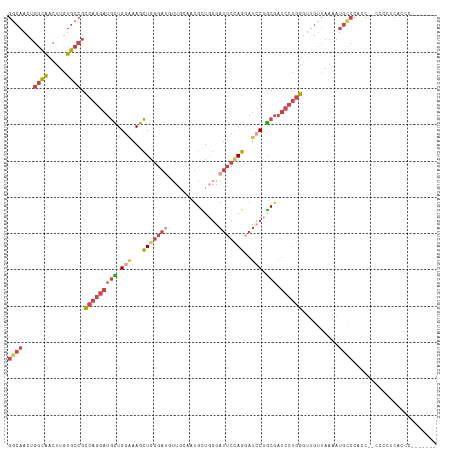
| Location | 7,223,418 – 7,223,515 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.96 |
| Shannon entropy | 0.14922 |
| G+C content | 0.54123 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -27.50 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7223418 97 + 24543557 GGAUCGCAGGAUACUAGAAUCCCAGCAUUGCAACAUCCCAGCUUUCCAACAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGU ((.((((.((((......))))((((((((((((...((((............)))).((((.....)))).)))).))))))))))))))...... ( -34.00, z-score = -2.93, R) >droSim1.chr3L 6734943 97 + 22553184 GGAUCGCAGGAUCCUAGAAUCCCAGCAUUGCAACAUCCCAGCUUUCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGU (((((....)))))(((..(((((((((((((((......(((....)))....(((((((....))))))))))).))))))))).)))))..... ( -39.40, z-score = -3.97, R) >droSec1.super_2 7149657 97 + 7591821 GGAUCGCAGGAUCCUAGAAUCCCAGCAGUGCAACAUCCCAGCUUUCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGU (((((....)))))(((..((((((((..(((((......(((....)))....(((((((....))))))))))))...)))))).)))))..... ( -37.20, z-score = -3.06, R) >droYak2.chr3L 7823902 97 + 24197627 GGAUCGCUGGAUCCUGGAAUCCCAACAACCCAACAUCCCAGCUUCACAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGU ((((.((((((..((((....................))))..)).)))))))).((((((....))))))((((((((....))))))))...... ( -36.65, z-score = -3.36, R) >droEre2.scaffold_4784 21642356 89 - 25762168 GGAUCGCAGCAUCCUGGAAUCCCA--------ACAUCCCAGCUUCCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGU ((((..(((....)))..))))..--------...(((((((......(((..((((((((....)))))))).)))....))))).))........ ( -30.10, z-score = -1.69, R) >consensus GGAUCGCAGGAUCCUAGAAUCCCAGCAUUGCAACAUCCCAGCUUUCCAGCAUCCUGGCGGCACAAGUUGCCAGUUGCCAAUGCUGGCGACUAAUAGU (((((....))))).....................(((((((......(((..((((((((....)))))))).)))....))))).))........ (-27.50 = -28.10 + 0.60)

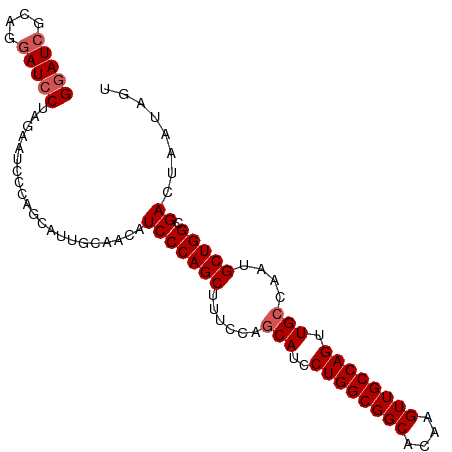
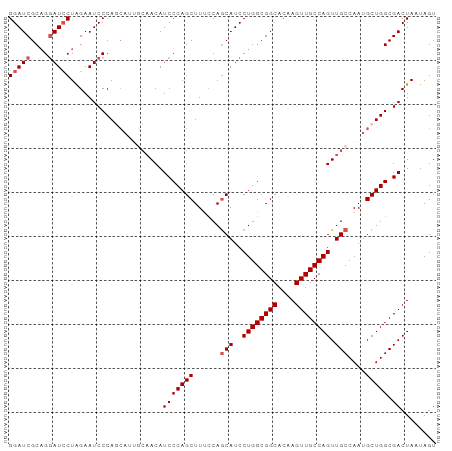
| Location | 7,223,418 – 7,223,515 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.96 |
| Shannon entropy | 0.14922 |
| G+C content | 0.54123 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -29.96 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7223418 97 - 24543557 ACUAUUAGUCGCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGUUGGAAAGCUGGGAUGUUGCAAUGCUGGGAUUCUAGUAUCCUGCGAUCC .......(((((((((((((.((((.((((.....)))).((((............))))...))))))))))))((((......)))).))))).. ( -34.80, z-score = -1.58, R) >droSim1.chr3L 6734943 97 - 22553184 ACUAUUAGUCGCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGAAAGCUGGGAUGUUGCAAUGCUGGGAUUCUAGGAUCCUGCGAUCC .......(((((((((((((.((((.((((.....)))).(((....(((....))))))...))))))))))))(((((....))))).))))).. ( -38.70, z-score = -2.30, R) >droSec1.super_2 7149657 97 - 7591821 ACUAUUAGUCGCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGAAAGCUGGGAUGUUGCACUGCUGGGAUUCUAGGAUCCUGCGAUCC .......((((((((((...(((((.((((.....)))).(((....(((....))))))...)))))..)))))(((((....))))).))))).. ( -36.50, z-score = -1.57, R) >droYak2.chr3L 7823902 97 - 24197627 ACUAUUAGUCGCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGUGAAGCUGGGAUGUUGGGUUGUUGGGAUUCCAGGAUCCAGCGAUCC .......(((.(((((..(((((.(((((..........)))))..)))))....))))))))...((((((((((.(((....))))))))))))) ( -37.60, z-score = -1.62, R) >droEre2.scaffold_4784 21642356 89 + 25762168 ACUAUUAGUCGCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGGAAGCUGGGAUGU--------UGGGAUUCCAGGAUGCUGCGAUCC .......(((((.((((((((((...(....)...)))).((((....))))....(((((((..--------....)))))))))))))))))).. ( -35.30, z-score = -1.72, R) >consensus ACUAUUAGUCGCCAGCAUUGGCAACUGGCAACUUGUGCCGCCAGGAUGCUGGAAAGCUGGGAUGUUGCAAUGCUGGGAUUCUAGGAUCCUGCGAUCC .......(((.(((((...((((...(....)...)))).((((....))))...))))))))............(((((.(((....))).))))) (-29.96 = -29.96 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:15 2011