
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,194,055 – 7,194,184 |
| Length | 129 |
| Max. P | 0.555314 |

| Location | 7,194,055 – 7,194,184 |
|---|---|
| Length | 129 |
| Sequences | 10 |
| Columns | 136 |
| Reading direction | forward |
| Mean pairwise identity | 78.05 |
| Shannon entropy | 0.48166 |
| G+C content | 0.46496 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7194055 129 + 24543557 -----CUACCAUUCCAUUCCAUUCCC-CUUCCCAAUCCCAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAAGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUCUAGUGGCUCAA-UGGGAGCGUUG -----.....................-.((((((....((((.......)))).....(((.((((((((((((((((((.....)))))).............))))))))....))))))).-))))))..... ( -29.31, z-score = -0.65, R) >droSim1.chr3L 6702248 133 + 22553184 CUACCAUUCCAUUCCAUACCAUUCCC-UU-CCCAAUCCCAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAAGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUCUAGUGGCUCAA-UGGGAGCAUUU ..........................-((-((((....((((.......)))).....(((.((((((((((((((((((.....)))))).............))))))))....))))))).-))))))..... ( -29.31, z-score = -0.61, R) >droSec1.super_2 7120495 134 + 7591821 CUACCAUUCCAUUUCAUACCAUUCCC-UUUCCCAAUCCCAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAAGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUCUAGUGGCUCAA-UGGGAGCAUUG ..........................-.((((((....((((.......)))).....(((.((((((((((((((((((.....)))))).............))))))))....))))))).-))))))..... ( -29.41, z-score = -0.58, R) >droYak2.chr3L 7792361 124 + 24197627 -----------UGGAAACCCGUUCCCGCUUCCCAAUCCCAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAAGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUCUAGUGGCUCAA-UGGGAGCGUUG -----------.((((.....))))((((.((((....((((.......)))).....(((.((((((((((((((((((.....)))))).............))))))))....))))))).-))))))))... ( -35.71, z-score = -1.42, R) >droEre2.scaffold_4784 21608839 126 - 25762168 --------UUUCCCAUUCCCGCUCCCGAU-CCCAAUCCCAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAAGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUCUAGUGGCUCAA-UGGGAGCGUUG --------...........((((((((..-........((((.......)))).....(((.((((((((((((((((((.....)))))).............))))))))....))))))).-))))))))... ( -36.71, z-score = -2.56, R) >droAna3.scaffold_13337 1721075 115 + 23293914 ------------------CUACUCC--CUGCCGACUCCCUGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAAGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUGUAGUGGCUCAAAUGGGGGAGCU- ------------------...((((--((........((..(((((...(((((((((((.(((.....)))..((((((.....)))))).....))))..))))))).)))))..))........))))))..- ( -31.29, z-score = 0.05, R) >dp4.chrXR_group6 3952331 128 + 13314419 -----CUUCUGUCCCACUUCACUCCG-CUGCCGA-UGCCAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAGGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUGUAGUGGCUCAA-UGGAAGGACUG -----.....((((........((((-.((....-.((((.(((((...(((((((((((.(((.....)))..((((((.....)))))).....))))..))))))).))))).)))).)).-)))).)))).. ( -35.20, z-score = -0.14, R) >droPer1.super_9 2241338 128 + 3637205 -----CUCCUGUCCCACUUCACUCCG-CUGCCGA-UGCCAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAGGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUGUAGUGGCUCAA-UGGAAGGACUG -----.....((((........((((-.((....-.((((.(((((...(((((((((((.(((.....)))..((((((.....)))))).....))))..))))))).))))).)))).)).-)))).)))).. ( -35.20, z-score = -0.14, R) >droWil1.scaffold_180698 7785537 119 + 11422946 --------AUAUAUAGGGGUCUUCUCUUUGCC--ACUCUAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAAGCCGGUUGCUUGAUUAACCAUUCAUUGCAGUUUUGUAGUGGCUCAA-UGGAA------ --------......(((((....))))).(((--(((.....((((...(((((((((((.(((.....)))..((((((.....)))))).....))))..))))))).))))))))))....-.....------ ( -29.40, z-score = -0.07, R) >droGri2.scaffold_15110 12843038 120 + 24565398 ------CUGCAGUUACUGUUACUGCCCCCCUUCUACUCGAAUGAGUGUAACGGCAAAAUUAAGCCAAAAGCAGUUGAAGCCGGCUGCUUGAUUUACCAUUCAUUGCUGUUUUAUAGUGCCUCAA-UG--------- ------..(((((.......)))))..............((((((((....(((........)))..(((((((((....))))))))).......))))))))(((((...))))).......-..--------- ( -30.20, z-score = -1.69, R) >consensus ______U_CCAUUCCAUUCCACUCCC_CUGCCCAAUCCCAGUGCAACUUGCUGUAAAAUGAAGCCAAAAGCUGUUCAAGCCGGCUGCUUGAUUAACCAUUCAUUGCAGUUUUCUAGUGGCUCAA_UGGGAGCAUUG ...................................(((((.((((......))))...(((.((((((((((((((((((.....)))))).............))))))))....)))))))..)))))...... (-21.60 = -22.20 + 0.60)

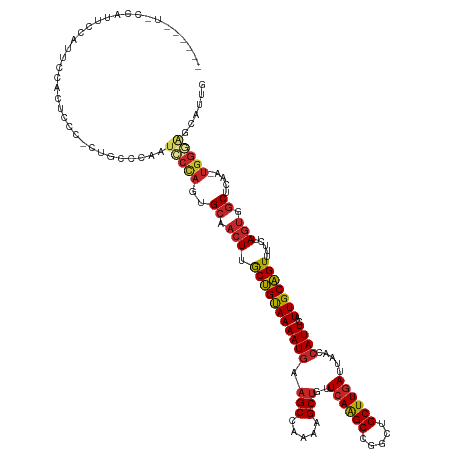
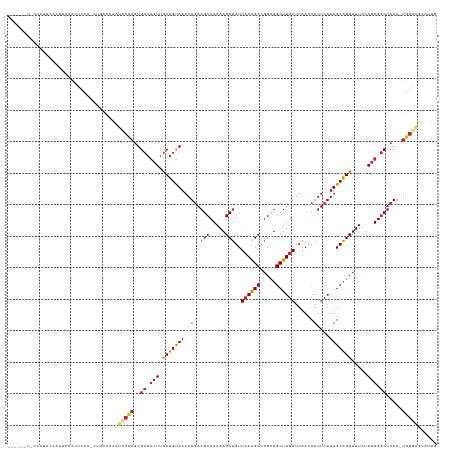
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:10 2011