| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,191,493 – 7,191,560 |
| Length | 67 |
| Max. P | 0.971254 |

| Location | 7,191,493 – 7,191,560 |
|---|---|
| Length | 67 |
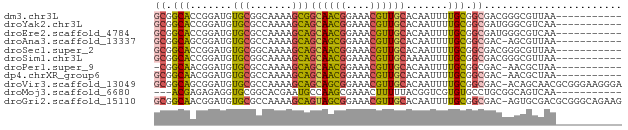
| Sequences | 11 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Shannon entropy | 0.42431 |
| G+C content | 0.57640 |
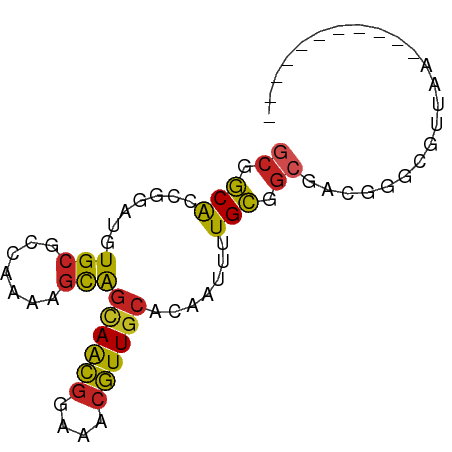
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -18.98 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971254 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
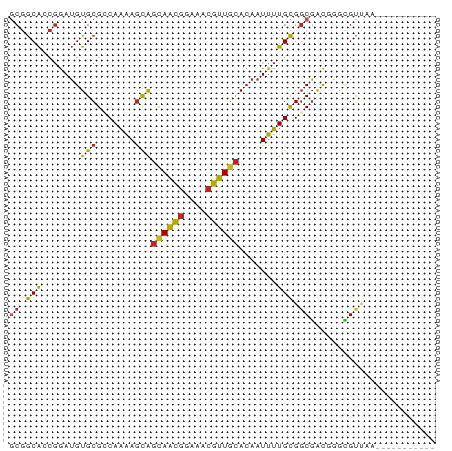
>dm3.chr3L 7191493 67 - 24543557 GCGGCACCGGAUGUGCGGCAAAAGCGGCAACGGAAACGUUGCACAAUUUUGCGGCGACGGGCGUUAA----------- ...((.(((....(((.((((((...((((((....))))))....)))))).))).))))).....----------- ( -32.50, z-score = -3.89, R) >droYak2.chr3L 7789746 67 - 24197627 GCGGCACCGGAUGUGCGCCAAAAGCAGCAACGGAAACGUUGCACAAUUUUGCGGCGAUGGGCGUCAA----------- ..(((.((.(.(((.(((.((((...((((((....))))))....)))))))))).).)).)))..----------- ( -25.90, z-score = -1.26, R) >droEre2.scaffold_4784 21606296 67 + 25762168 GCGGCACCGGAUGUGCGCCAAAAGCAGCAACGGAAACGUUGCACAAUUUUGCGGCGAUGGGCGUCAA----------- ..(((.((.(.(((.(((.((((...((((((....))))))....)))))))))).).)).)))..----------- ( -25.90, z-score = -1.26, R) >droAna3.scaffold_13337 1718539 66 - 23293914 GCGGCAGCGGAUGUGCGCCAAAAGCAGCAGCGGAAACGUUGCACAAUUUUGCGGCGAC-AGCGUUAA----------- ((....)).(((((.((((.((((..((((((....))))))....))))..))))..-.)))))..----------- ( -24.90, z-score = -0.87, R) >droSec1.super_2 7117611 67 - 7591821 GCGGCACCGGAUGUGCGGCAAAAGCAGCAACGGAAACGUUGCACAAUUUUGCGGCGACGGGCGUUAA----------- ...((.(((....(((.((((((...((((((....))))))....)))))).))).))))).....----------- ( -31.40, z-score = -3.79, R) >droSim1.chr3L 6699412 67 - 22553184 GCGGCACCGGAUGUGCGGCAAAAGCAGCAACGGAAACGUUGCAAAAUUUUGCGGCGACGGGCGUUAA----------- ...((.(((....(((.((((((...((((((....))))))....)))))).))).))))).....----------- ( -31.40, z-score = -3.92, R) >droPer1.super_9 2238708 65 - 3637205 -CGGCAACGGAUGUGCGCCAAAAGCAGCAACGGAAACGUUGCACAAUUUUGCGGCGAC-AACGCUAA----------- -((....))..(((.((((.((((..((((((....))))))....))))..))))))-).......----------- ( -23.70, z-score = -2.12, R) >dp4.chrXR_group6 3949746 66 - 13314419 GCGGCAACGGAUGUGCGCCAAAAGCAGCAACGGAAACGUUGCACAAUUUUGCGGCGAC-AACGCUAA----------- (((....)...(((.((((.((((..((((((....))))))....))))..))))))-)..))...----------- ( -24.00, z-score = -1.76, R) >droVir3.scaffold_13049 9430392 77 + 25233164 GCGGCAGCGGAUGUGCGCCAAAAGCAGCAGCGGAAACGUUGCACAAUUUUGCGGCGAC-ACAGCAACGCGGGAAGGGA ......(((..((((((((.((((..((((((....))))))....))))..)))).)-)))....)))......... ( -29.40, z-score = -1.82, R) >droMoj3.scaffold_6680 3744524 64 + 24764193 ---ACGAGAGAGGUGCGGCACGAAUGCCAAGCGAAACUUUUUACGGUCGUGUGCCUGCGGCAGUCAA----------- ---.((((((((.(((((((....))))..)))...)))))).))(((((......)))))......----------- ( -18.70, z-score = 0.32, R) >droGri2.scaffold_15110 12840471 77 - 24565398 GCGGCAACGGAUGUGCGCCAAAAGCAGUAGCGGAAACGUUGCACAAUUUUGCGGCGAC-AGUGCGACGCGGGCAGAAG .((....))..(.(((.((.......((((((....))))))........(((.((..-....)).)))))))).).. ( -26.00, z-score = -0.27, R) >consensus GCGGCACCGGAUGUGCGCCAAAAGCAGCAACGGAAACGUUGCACAAUUUUGCGGCGACGGGCGUUAA___________ ((.(((.......(((.......)))((((((....)))))).......))).))....................... (-18.98 = -18.70 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:09 2011