
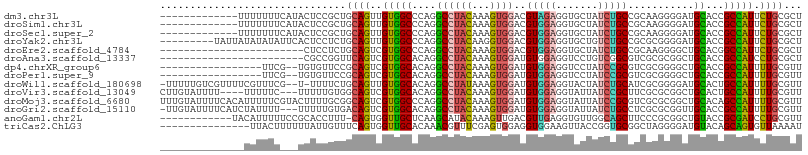
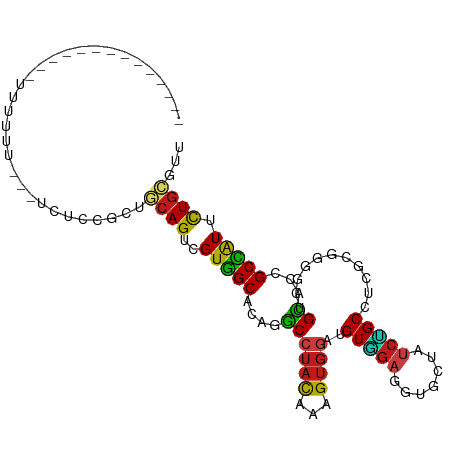
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,176,567 – 7,176,663 |
| Length | 96 |
| Max. P | 0.877989 |

| Location | 7,176,567 – 7,176,661 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 68.60 |
| Shannon entropy | 0.67348 |
| G+C content | 0.54522 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -15.06 |
| Energy contribution | -14.59 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7176567 94 - 24543557 -------------UUUUUUUCAUACUCCGCUGCAGUUGUGGCCCAGGCCUACAAAGUGGACGUAGAGGUGCUAUCUGCCGCAAGGGGAUGCACCGCCAUUCUGCGCU -------------............((((((....(((((((....))).))))))))))(((((((((((.((((.((....))))))))))).....)))))).. ( -37.00, z-score = -2.01, R) >droSim1.chr3L 6682631 94 - 22553184 -------------UUUUUUUCAUACUCCGCUGCAGUUGUGGCCCAGGCCUACAAAGUGGACGUGGAGGUGCUAUCUGCCGCAAGGGGAUGCACCGCCAUUCUGCGCU -------------...............((.((((..((((((((.(((........)).).))).(((((.((((.((....)))))))))))))))).)))))). ( -40.30, z-score = -2.59, R) >droSec1.super_2 7102639 94 - 7591821 -------------UUUUUUUCAUACUCCGCUGCAGUUGUGGCCCAGGCCUACAAAGUGGACGUGGAGGUGCUAUCUGCCGCAAGGGGAUGCACCGCCAUUCUGCGCU -------------...............((.((((..((((((((.(((........)).).))).(((((.((((.((....)))))))))))))))).)))))). ( -40.30, z-score = -2.59, R) >droYak2.chr3L 7774449 98 - 24197627 ---------UAUUAUAUAUAUUCACUCCUCUGCAGUUGUGGCCCAGGCCUACAAGGUGGACGUGGAGGUGCUGUCUGCCGCGCGGGGAUGCACCGCCAUUCUGCGCU ---------......................((((..((((((((.(((........)).).))).((((((.(((((...))))).).)))))))))).))))... ( -35.40, z-score = -0.23, R) >droEre2.scaffold_4784 21591298 83 + 25762168 ------------------------CUCCUCUGCAGUCGUGGCCCAGGCCUACAAAGUGGACGUGGAGGUGCUAUCUGCCGCAAGGGGCUGCACGGCCAUUCUGCGCU ------------------------.......((((..((((((((.(((........)).).))...((((...((.((....))))..)))))))))).))))... ( -31.30, z-score = 0.46, R) >droAna3.scaffold_13337 1704658 83 - 23293914 ------------------------CGCCGGUUCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUCCUGUCGGCGUCGCGCGGCUGCACCGCCAUCCUGCGCU ------------------------.((.(((.((((((((.((((..((........)).))))((.(((.....))).)).)))))))).)))((......)))). ( -30.80, z-score = 1.15, R) >dp4.chrXR_group6 3934147 88 - 13314419 -----------------UUCG--UGUGUUCCGCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUCCUAUCCGCGUCGCGGGGCUGCACCGCCAUUUUGCGUU -----------------...(--((((((((((....(((((....))).))...(((((((.((....)))))))))...))))))).))))(((......))).. ( -35.30, z-score = -1.27, R) >droPer1.super_9 2223187 88 - 3637205 -----------------UUCG--UGUGUUCCGCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUCCUAUCCGCGUCGCGGGGCUGCACCGCCAUUUUGCGUU -----------------...(--((((((((((....(((((....))).))...(((((((.((....)))))))))...))))))).))))(((......))).. ( -35.30, z-score = -1.27, R) >droWil1.scaffold_180698 7762209 103 - 11422946 -UUUUUGUCGUUUUCGUUUCG--U-UUUUCUGCAGUUGUGGCACAGGCCUAUAAAGUGGAUGUGGAGGUACUAUCUGCAUCGCGGGGAUGCACUGCCAUUUUGCGUU -....................--.-......((((..((((((((..(((.....(((.(((..((.......))..)))))).))).))...)))))).))))... ( -25.20, z-score = 0.53, R) >droVir3.scaffold_13049 9415062 100 + 25233164 CUUGUAUUUU----UUUUUC---UUUUUGUGGCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUAUUAUCCGCUUCGCGCGGCUGCACUGCCAUUUUGCGUU ..........----......---.....((((((((.(..((...(.....)...(((((.(((((.......))))))))))...))..)))))))))........ ( -30.00, z-score = -0.89, R) >droMoj3.scaffold_6680 20993941 107 - 24764193 UUUGUAUUUUCACAUUUUUCGUACUUUUGCGGCAGUCGUGGCCCAGGCCUACAAAGUGGAUGUGGAGGUAUUAUCCGCGUCGCGCGGCUGCACAGCCAUUUUGCGUU ...................((((.....((.(((((((((((....))).))...(((((((((((.......)))))))).)))))))))...)).....)))).. ( -37.10, z-score = -1.93, R) >droGri2.scaffold_15110 12824674 103 - 24565398 -UUGUAUUUUCAUCUAUUUU---UUUUUGUGACAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUAUUAUCUGCCUCGCGCGGUUGCACCGCCAUUUUGCGUU -...................---.....(((.((..((((.((((..((........)).))))((((((.....)))))).))))..)))))(((......))).. ( -28.30, z-score = -0.71, R) >anoGam1.chr2L 43587874 94 + 48795086 ------------UACAUUUUUCCGCACCUUU-CAGUGGUUGCUCAAGCAUACAAAGUUGACGUUGAGGUGUUGGCAGCUUCCCGCGGCUGUACCGCGAUCCUGCGUU ------------.........(.((((((((-(((((..(((....)))..))...))))....))))))).)((((.....(((((.....)))))...))))... ( -26.00, z-score = -0.01, R) >triCas2.ChLG3 6241126 92 + 32080666 ---------------UUACUUUUUUAUUGUUUCAGUGGUUGCACAAACGUUUCGAGUGGAGGUGGAAGUUACCGGUGCGGCUAGGGGAUGUACAGCAGUGUUAAAAU ---------------....(((..(((((((((..(((((((((..((.......))...((((.....)))).)))))))))..)))......))))))..))).. ( -20.20, z-score = 0.03, R) >consensus ______________UUUUUU___UCUCCGCUGCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUGCUAUCUGCCUCGCGGGGCUGCACCGCCAUUCUGCGUU ...............................((((..(((((....((((((...))))..(((((.......)))))...........))...))))).))))... (-15.06 = -14.59 + -0.46)

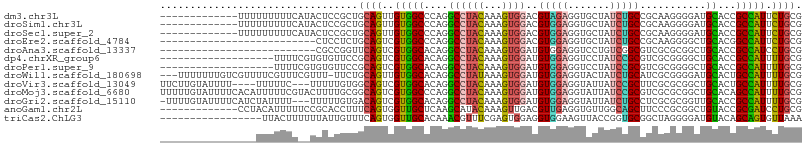
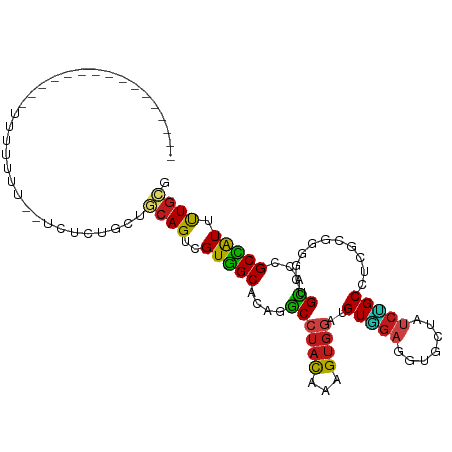
| Location | 7,176,569 – 7,176,663 |
|---|---|
| Length | 94 |
| Sequences | 13 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.69356 |
| G+C content | 0.54373 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.28 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7176569 94 - 24543557 -------------UUUUUUUUUCAUACUCCGCUGCAGUUGUGGCCCAGGCCUACAAAGUGGACGUAGAGGUGCUAUCUGCCGCAAGGGGAUGCACCGCCAUUCUGCG -------------..............((((((....(((((((....))).))))))))))(((((((((((.((((.((....))))))))))).....)))))) ( -36.60, z-score = -2.48, R) >droSim1.chr3L 6682633 94 - 22553184 -------------UUUUUUUUUCAUACUCCGCUGCAGUUGUGGCCCAGGCCUACAAAGUGGACGUGGAGGUGCUAUCUGCCGCAAGGGGAUGCACCGCCAUUCUGCG -------------....................((((..((((((((.(((........)).).))).(((((.((((.((....)))))))))))))))).)))). ( -38.50, z-score = -2.60, R) >droSec1.super_2 7102641 94 - 7591821 -------------UUUUUUUUUCAUACUCCGCUGCAGUUGUGGCCCAGGCCUACAAAGUGGACGUGGAGGUGCUAUCUGCCGCAAGGGGAUGCACCGCCAUUCUGCG -------------....................((((..((((((((.(((........)).).))).(((((.((((.((....)))))))))))))))).)))). ( -38.50, z-score = -2.60, R) >droEre2.scaffold_4784 21591300 81 + 25762168 --------------------------CUCCUCUGCAGUCGUGGCCCAGGCCUACAAAGUGGACGUGGAGGUGCUAUCUGCCGCAAGGGGCUGCACGGCCAUUCUGCG --------------------------.......((((..((((((((.(((........)).).))...((((...((.((....))))..)))))))))).)))). ( -31.30, z-score = -0.04, R) >droAna3.scaffold_13337 1704660 81 - 23293914 --------------------------CGCCGGUUCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUCCUGUCGGCGUCGCGCGGCUGCACCGCCAUCCUGCG --------------------------.(.((((.((((((((.((((..((........)).))))((.(((.....))).)).)))))))).)))).)........ ( -30.60, z-score = 0.68, R) >dp4.chrXR_group6 3934149 88 - 13314419 -------------------UUUUCGUGUGUUCCGCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUCCUAUCCGCGUCGCGGGGCUGCACCGCCAUUUUGCG -------------------.....(((((((((((....(((((....))).))...(((((((.((....)))))))))...))))))).))))(((......))) ( -34.70, z-score = -1.34, R) >droPer1.super_9 2223189 88 - 3637205 -------------------UUUUCGUGUGUUCCGCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUCCUAUCCGCGUCGCGGGGCUGCACCGCCAUUUUGCG -------------------.....(((((((((((....(((((....))).))...(((((((.((....)))))))))...))))))).))))(((......))) ( -34.70, z-score = -1.34, R) >droWil1.scaffold_180698 7762211 103 - 11422946 ---UUUUUUUGUCGUUUUCGUUUCGUUU-UUCUGCAGUUGUGGCACAGGCCUAUAAAGUGGAUGUGGAGGUACUAUCUGCAUCGCGGGGAUGCACUGCCAUUUUGCG ---.........................-....((((..((((((((..(((.....(((.(((..((.......))..)))))).))).))...)))))).)))). ( -25.20, z-score = 0.29, R) >droVir3.scaffold_13049 9415064 100 + 25233164 UUCUUGUAUUUU----UUUUUC---UUUUUGUGGCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUAUUAUCCGCUUCGCGCGGCUGCACUGCCAUUUUGCG ............----......---.....((((((((.(..((...(.....)...(((((.(((((.......))))))))))...))..)))))))))...... ( -30.00, z-score = -1.14, R) >droMoj3.scaffold_6680 20993943 107 - 24764193 UUUUUGUAUUUUCACAUUUUUCGUACUUUUGCGGCAGUCGUGGCCCAGGCCUACAAAGUGGAUGUGGAGGUAUUAUCCGCGUCGCGCGGCUGCACAGCCAUUUUGCG .....................((((.....((.(((((((((((....))).))...(((((((((((.......)))))))).)))))))))...)).....)))) ( -36.50, z-score = -2.05, R) >droGri2.scaffold_15110 12824676 103 - 24565398 -UUUUGUAUUUUCAUCUAUUUU---UUUUUGUGACAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUAUUAUCUGCCUCGCGCGGUUGCACCGCCAUUUUGCG -.....................---.....(((.((..((((.((((..((........)).))))((((((.....)))))).))))..)))))(((......))) ( -27.70, z-score = -0.81, R) >anoGam1.chr2L 43587876 94 + 48795086 -------------CCUACAUUUUUCCGCACCUUUCAGUGGUUGCUCAAGCAUACAAAGUUGACGUUGAGGUGUUGGCAGCUUCCCGCGGCUGUACCGCGAUCCUGCG -------------.(.(((((((..((((.((((..(((..(((....)))))))))).)).))..))))))).)((((.....(((((.....)))))...)))). ( -26.20, z-score = -0.24, R) >triCas2.ChLG3 6241128 90 + 32080666 -----------------UUACUUUUUUAUUGUUUCAGUGGUUGCACAAACGUUUCGAGUGGAGGUGGAAGUUACCGGUGCGGCUAGGGGAUGUACAGCAGUGUUAAA -----------------.((((.((.(((...(((..(((((((((..((.......))...((((.....)))).)))))))))..))).))).)).))))..... ( -19.40, z-score = 0.36, R) >consensus ________________UUUUUUU__UCUCUGCUGCAGUCGUGGCACAGGCCUACAAAGUGGAUGUGGAGGUGCUAUCUGCCUCGCGGGGCUGCACCGCCAUUUUGCG .................................((((..(((((....((((((...))))..(((((.......)))))...........))...))))).)))). (-14.74 = -14.28 + -0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:07 2011