| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,175,739 – 7,175,825 |
| Length | 86 |
| Max. P | 0.654198 |

| Location | 7,175,739 – 7,175,825 |
|---|---|
| Length | 86 |
| Sequences | 9 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 69.88 |
| Shannon entropy | 0.63184 |
| G+C content | 0.48452 |
| Mean single sequence MFE | -15.65 |
| Consensus MFE | -7.46 |
| Energy contribution | -7.39 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
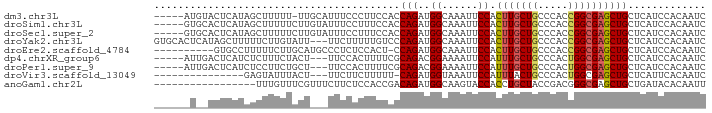
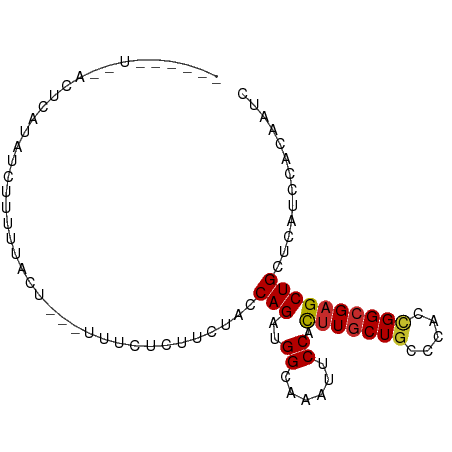
>dm3.chr3L 7175739 86 - 24543557 -----AUGUACUCAUAGCUUUUU-UUGCAUUUCCCUUCCACCAGAUGGCAAAUUCCACUUGCUGCCCACCGGCGAGCUGCUCAUCCACAAUC -----.(((.......((.....-..))...............(((((((.......(((((((.....))))))).))).)))).)))... ( -14.60, z-score = -0.34, R) >droSim1.chr3L 6681833 87 - 22553184 -----GUGCACUCAUAGCUUUUUCUUGUAUUUCCUUUCCACCAGAUGGCAAAUUCCACUUGCUGCCCACCGGCGAGCUGCUCAUCCACAAUC -----(.((((((...(((...(((.((...........)).))).)))..............(((....)))))).))).).......... ( -15.50, z-score = -0.55, R) >droSec1.super_2 7101846 87 - 7591821 -----GUGCACUCAUAGCUUUUUCUUGUAUUUCCUUUCCACCAGAUGGCAAAUUCCACUUGCUGCCCACCGGCGAGCUGCUCAUCCACAAUC -----(.((((((...(((...(((.((...........)).))).)))..............(((....)))))).))).).......... ( -15.50, z-score = -0.55, R) >droYak2.chr3L 7773608 89 - 24197627 GUGCACUCAUAGCUUUUUCUUGUAUU---UUCUUUUUGUCCCAGAUGGCAAAUUCCACUUGCUGCCCACCGGCGAGCUGCUCAUCCACAAUC (.((((((.............(((..---.....((((((......)))))).......))).(((....)))))).))).).......... ( -16.64, z-score = -0.57, R) >droEre2.scaffold_4784 21590501 81 + 25762168 ----------GUGCCUUUUUCUUGCAUGCCCUCUCCACU-CCAGAUGGCAAAUUCCACUUGCUGCCCACCGGCGAGCUGCUCAUCCACAAUC ----------(((.............((((.(((.....-..))).)))).....(((((((((.....))))))).))......))).... ( -17.10, z-score = -0.87, R) >dp4.chrXR_group6 3933387 84 - 13314419 -----AUUGACUCAUCUCUUUCUACU---UUCCACUUUUCGCAGACGGAAAAUUCCAUUUGCUGCCCACUGGCGAGCUGCUCAUCCACAAUC -----..(((..((.(((........---...........(((((.(((....))).))))).(((....)))))).)).)))......... ( -16.00, z-score = -1.96, R) >droPer1.super_9 2222426 84 - 3637205 -----AUUGACUCAUCUCCUUCUGCU---UUCCACUUUUCGCAGACGGAAAAUUCCAUUUGCUGCCCACUGGCGAGCUGCUCAUCCACAAUC -----((((.......(((.(((((.---...........))))).)))...........((((((....))).)))..........)))). ( -17.30, z-score = -1.65, R) >droVir3.scaffold_13049 9414129 73 + 25233164 ---------------GAGUAUUUACU---UUCUUCUUUUU-CAGAUGGUAAAUUCCAUUUACUGCCCACUGGCGAGCUGCUCAUUCACAAUC ---------------(((((((((((---.(((.......-.))).))))))..........((((....))))...))))).......... ( -14.10, z-score = -1.89, R) >anoGam1.chr2L 43587659 75 + 48795086 -----------------UUUGUUUCGUUUCUUCUCCACCGACAGAUGGCAAGUACCACCUGCUACCGACGGGCGAGCUGCUGAUACACAAUU -----------------..(((.(((...(..(((..(((...(.(((((.(.....).))))).)..)))..)))..).))).)))..... ( -14.10, z-score = 0.39, R) >consensus ______U__ACUCAUAUCUUUUUACU___UUUCUCUUCUACCAGAUGGCAAAUUCCACUUGCUGCCCACCGGCGAGCUGCUCAUCCACAAUC .........................................(((..((......)).(((((((.....))))))))))............. ( -7.46 = -7.39 + -0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:06 2011