
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,170,729 – 7,170,832 |
| Length | 103 |
| Max. P | 0.672007 |

| Location | 7,170,729 – 7,170,832 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Shannon entropy | 0.42057 |
| G+C content | 0.52193 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7170729 103 + 24543557 GAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCUCGAGGCGUCUGCCCCCCAUCCACCAUCCA-CCAUCUAC------ ....(((...((((((((((.((((.((......)))))).))))....))))))......((((....))))...)))................-........------ ( -24.10, z-score = -0.76, R) >droSim1.chr3L 6676760 103 + 22553184 AAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCUCGAGGCGUCUGCCCCCCAUCCACCAUCCA-GCACCUAC------ ....(((...((((((((((.((((.((......)))))).))))....))))))......((((....))))...)))................-........------ ( -24.10, z-score = -0.58, R) >droSec1.super_2 7096836 96 + 7591821 AAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCGUUUAAUUUCCGCCUCGAGGCGUCUGCCCCCCAUCCAGCACCUA-G------------- ....((((.(((.((((((.(((.((((.(((......))))))).))).)))))).))).))))...(((.(.(((.........)))).))).-.------------- ( -27.00, z-score = -1.46, R) >droYak2.chr3L 7768416 100 + 24197627 AAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCUCGAGGCGUCUGCCCUCCAUCCACUUUCCA-CCACC--------- ....(((...((((((((((.((((.((......)))))).))))....))))))......((((....))))...)))................-.....--------- ( -24.10, z-score = -1.01, R) >droAna3.scaffold_13337 1699728 80 + 23293914 AAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCCCAAGGCGUCUACAG------------------------------ ....(((((.....(.(((.((((((((((((((((...))).)))))).)))))))....)))).....))))).....------------------------------ ( -21.20, z-score = -0.87, R) >dp4.chrXR_group6 3928754 96 + 13314419 AAAUGGCGUAAAAUGAGCGUUUGAGUGCUGAGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCCCAAGGCUCCUCCUUGCCGUCUGGCUCC---UCG----------- ....((((.(((.((((.(.(((.((((((((....)))).)))).))).).)))).))).))))(((.(((........)))...)))....---...----------- ( -26.60, z-score = -1.45, R) >droPer1.super_9 2217567 96 + 3637205 AAAUGGCGUAAAAUGAGCGUUUGAGUGCUGAGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCCCAAGGCUCCUCCUUGCCGUCUGGCUCC---UCG----------- ....((((.(((.((((.(.(((.((((((((....)))).)))).))).).)))).))).))))(((.(((........)))...)))....---...----------- ( -26.60, z-score = -1.45, R) >droWil1.scaffold_180698 7756569 107 + 11422946 AAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCCCAAGGCGUCUACUGCUCCACAUCCUUCCA-UCCUUCCAUCCA-- ..((((........((((..((((((((((((((((...))).)))))).)))))))....((((....))))......)))).........)))-)...........-- ( -24.73, z-score = -1.76, R) >droVir3.scaffold_13049 9409927 96 - 25233164 AAAUGGCGUAAAAUGAGCGUUUGAGUGUUACGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCCCAAGGCGUCUACUUCAGCUCCGGCUCC---UCG----------- ....((.((.....(((((.(((.((((.(((......))))))).))).)..........((((....)))).........))))..)).))---...----------- ( -22.70, z-score = -0.22, R) >droMoj3.scaffold_6680 20986388 110 + 24764193 AAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCCCAAGGCGUCAGCUCCAGCGUCAGCUUCAGCUCGGCUCCUGCUCG ..............((((..((((((((((((((((...))).)))))).)))))))...........(((.(((((((..(((....)))..)))).))).))))))). ( -33.30, z-score = -0.83, R) >droGri2.scaffold_15110 12819609 96 + 24565398 AAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCCCAAGGCAUCUGCUCCAGCUCCAGUUCC---UCG----------- ...(((.((.....((((..((((((((((((((((...))).)))))).))))))).....(((....)))....))))..)).))).....---...----------- ( -24.70, z-score = -1.27, R) >consensus AAAUGGCGUAAAAUGAGCGUUUGAGUGUUGCGUUACCUCGUGCGCGCAAUCAUUUAAUUUCCGCCCCAAGGCGUCUGCUCCCCAUCCAGCUCC___UCG___________ ....((.((.((((((((((.((((.((......)))))).))))....)))))).))..))(((....)))...................................... (-18.18 = -18.28 + 0.10)

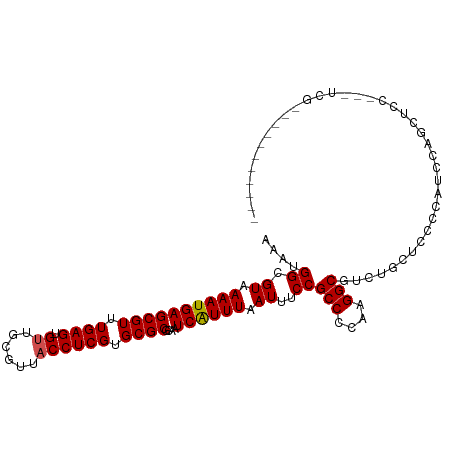

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:09:05 2011