| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,550,756 – 4,550,849 |
| Length | 93 |
| Max. P | 0.513711 |

| Location | 4,550,756 – 4,550,849 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Shannon entropy | 0.39253 |
| G+C content | 0.38006 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -13.37 |
| Energy contribution | -12.92 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4550756 93 - 23011544 UCUUAAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUC-----ACAUAUGCUUGAA-ACAAU-----UUGAAACCAAUUGA---- .........--..(((.((.(---(((((((.((((((((...((((....))))...))))))))-----.((......))..-.))))-----)))).))...))).---- ( -22.30, z-score = -2.15, R) >droSim1.chr2L_random 245131 93 - 909653 UCUUAAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUC-----ACAUAUGCUUGAA-ACAAU-----UUGAAACCAAUUGA---- .........--..(((.((.(---(((((((.((((((((...((((....))))...))))))))-----.((......))..-.))))-----)))).))...))).---- ( -22.30, z-score = -2.15, R) >droSec1.super_5 2643499 93 - 5866729 UCUUAAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUC-----ACAUAUGCUUGAA-ACAAU-----UUGAAACAAAUUGA---- .........--......((.(---(((((((.((((((((...((((....))))...))))))))-----.((......))..-.))))-----)))).)).......---- ( -23.00, z-score = -2.79, R) >droYak2.chr2L 4568157 93 - 22324452 UCUUAAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUC-----ACAUAUGCUUGAA-ACAAU-----UUCAAACCAAUUGA---- .........--..........---..((((((((((((((...((((....))))...))))))))-----........(((((-.....-----))))).))))))..---- ( -21.30, z-score = -1.97, R) >droEre2.scaffold_4929 4634196 93 - 26641161 UCUUAAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUC-----ACAUAUGCUUGAA-ACAAU-----UUGAAACCAAUUGA---- .........--..(((.((.(---(((((((.((((((((...((((....))))...))))))))-----.((......))..-.))))-----)))).))...))).---- ( -22.30, z-score = -2.15, R) >droAna3.scaffold_12943 1750054 94 + 5039921 UCUUAAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUC-----ACAUAUGCUUGAAUCCGAU-----UCGAAACCGAUCGA---- .........--..........---..((((((((((((((...((((....))))...))))))))-----........((((((...))-----))))..))))))..---- ( -23.40, z-score = -1.35, R) >dp4.chr4_group3 387988 97 + 11692001 UCUCAAAUGAAAAUAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUCACACAACAUAUGCUUGA----AAU-----UUGAAACCAAUUGA---- ..((((((.......((((..---....(((.((((((((...((((....))))...))))))))...))).....))))..----.))-----))))..........---- ( -19.40, z-score = -1.01, R) >droPer1.super_8 1434615 97 + 3966273 UCUCAAAUGAAAAUAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUCACACAACAUAUGCUUGA----AAU-----UUGAAACCAAUUGA---- ..((((((.......((((..---....(((.((((((((...((((....))))...))))))))...))).....))))..----.))-----))))..........---- ( -19.40, z-score = -1.01, R) >droWil1.scaffold_180703 1293569 88 + 3946847 UCUUGAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUCACAU-ACAUAUGCUUU-------------UUAAAAUCUAAU------ .........--...(((((..---.....((.((((((((...((((....))))...)))))))).)).-......)))))-------------............------ ( -16.24, z-score = -1.08, R) >droVir3.scaffold_12963 8461429 103 + 20206255 UCUCGAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUGGUUC-----ACAUAUGCUUAAAGUCAAUUUUUGUUAUUGUCAGUUCAAGUG ....((((.--.((((...(.---.((((((.((((((((...((((....))))...))))))))-----......((.....))))))))..)...))))..))))..... ( -20.80, z-score = 0.24, R) >droGri2.scaffold_15126 3101690 101 + 8399593 -CUUGAAUG--AACAAAGUCU---CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUGGUUC-----ACAUAUGCUUAAAAUCAAUAUUU-UUUUUGUCAGUUCAAGUG -(((((((.--.((((((...---...(((((((((((((...((((....))))...))))))))-----...((....))...)))))....-.))))))..))))))).. ( -24.30, z-score = -1.60, R) >anoGam1.chr3R 4723449 92 + 53272125 UUGUGUAUC-GGAUUGCUUCUCGCCAGAUUGGGAACUGUGGACGGGACACUUCUCACAAAUAGUUCACU--ACUUGCACGUGU------------UAGGCCUCGGGU------ ......(((-.((..((((..(((..((.((((((((((....((((....))))....))))))).))--).))....))).------------.)))).)).)))------ ( -20.90, z-score = 1.14, R) >consensus UCUUAAAUG__AACAAAGUCU___CAGAUUGGGAACUGUGGACGGGACAAUUCUCACACAUAGUUC_____ACAUAUGCUUGAA_ACAAU_____UUGAAACCAAUUGA____ ..........................((((((((((((((...((((....))))...))))))))........((....))...................))))))...... (-13.37 = -12.92 + -0.45)

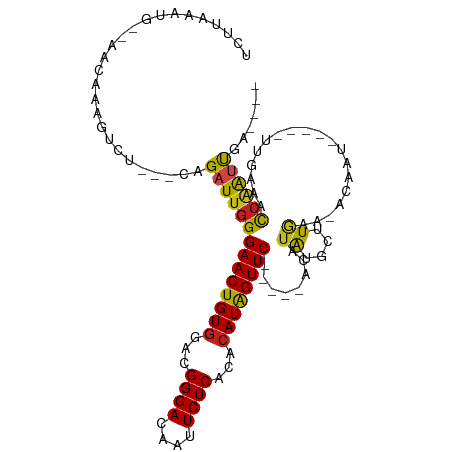

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:40 2011