
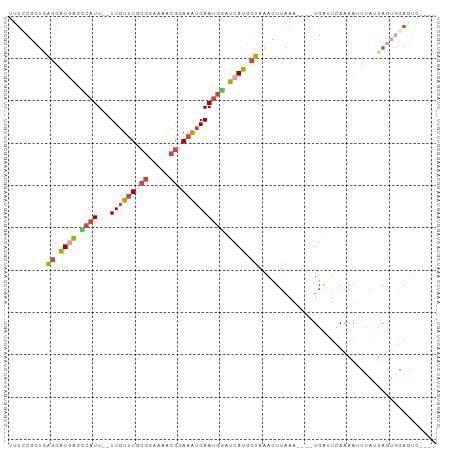
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 494,829 – 494,956 |
| Length | 127 |
| Max. P | 0.823531 |

| Location | 494,829 – 494,920 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 68.14 |
| Shannon entropy | 0.59135 |
| G+C content | 0.37025 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -8.80 |
| Energy contribution | -9.25 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 494829 91 - 23011544 UUUCCGCUGAGGAUGAGCCAUU--UUGUUUGCGGAAAACGGAAAUGAAUGGAUCAUGCUAAACUUAAA----UGAUUCAAAAUCUAUUAGUGGAGUC---- .(((((((((((((.......(--(..(((.((.....)).)))..))((((((((...........)----)))))))..)))).)))))))))..---- ( -25.90, z-score = -3.12, R) >droEre2.scaffold_4929 547161 92 - 26641161 ---UCGCUGAGGAUGAGCCAUU--UUGUUUGCGGAAGGCGGAAAUGAAUGAAUAAUGCUAGAUUUAAAUAAGUGAAUCAAGAAUAAAUAGUGGAGGC---- ---((((((((.((....((((--(..(((((.....)))))...)))))....)).)).((((((......)))))).........))))))....---- ( -17.30, z-score = -1.07, R) >droYak2.chr2L 479436 92 - 22324452 UUUCCGCUAAGGAUGAGGCAUU--UUGUUUGCGGAA-ACGGAAAUGAAUGCAUCAUGCUAAACUUAAG----UGAAUAAAGAGUCAACAAGUAAAUAAU-- .........((.((((.((((.--(..(((.((...-.)).)))..))))).)))).))..((((...----(((........)))..)))).......-- ( -21.60, z-score = -2.37, R) >droSec1.super_14 477602 91 - 2068291 UUUCCGCUGAGGAUGAGCCAUU--UUGUUUGCGGAAAACGGAAAUGAAUGGAUCAUGCUAAACUUAAA----UGAUUCAAAAUCUGUUAGUGGAGUC---- .(((((((((((((.......(--(..(((.((.....)).)))..))((((((((...........)----)))))))..)))).)))))))))..---- ( -25.90, z-score = -2.92, R) >droSim1.chr2L 501272 91 - 22036055 UUUCCGCUGAGGAUGAGCCAUU--UUGUUUGCGGAAAACGGAAAUGAAUGGAUCAUGCUAAACUUAAA----UGAUUCAAAAUCUAUUAGUAGAGUC---- .(((..((((((((.......(--(..(((.((.....)).)))..))((((((((...........)----)))))))..)))).))))..)))..---- ( -20.60, z-score = -1.52, R) >droPer1.super_1 2596519 94 - 10282868 UCUCCAUCGCUUCUUGAAAGUUGAUUGUAUCCCAAA---AGAAGGGAAGAUAUCAGAUGAAAAUAUAU----UGGCUUAUGGGGUUUCAUAGCACUGAUUC ........(((.........(((((....((((...---....))))....)))))((((((.((((.----.....))))...)))))))))........ ( -16.20, z-score = 1.12, R) >consensus UUUCCGCUGAGGAUGAGCCAUU__UUGUUUGCGGAAAACGGAAAUGAAUGGAUCAUGCUAAACUUAAA____UGAUUCAAAAUCUAUUAGUGGAGUC____ .........((.((((.((((...((((((.((.....)).)))))))))).)))).)).......................................... ( -8.80 = -9.25 + 0.45)


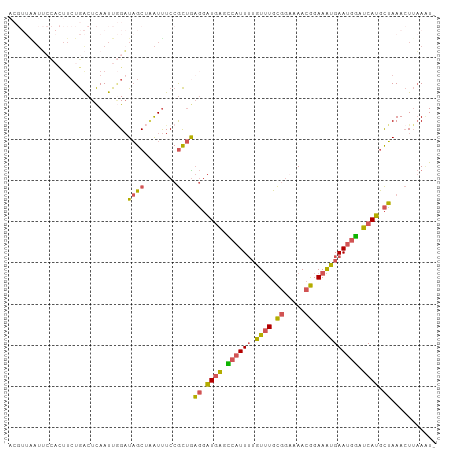
| Location | 494,853 – 494,956 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.87 |
| Shannon entropy | 0.54576 |
| G+C content | 0.37686 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -13.00 |
| Energy contribution | -12.32 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 494853 103 - 23011544 ACGUUAAUUCCACUUCUGACUCAAUUGGAUAGCUAAUUUCCGCUGAGGAUGAGCCAUUUUGUUUGCGGAAAACGGAAAUGAAUGGAUCAUGCUAAACUUAAAU- ..(((...((((.............))))((((........))))((.((((.((((.(..(((.((.....)).)))..))))).)))).)).)))......- ( -20.82, z-score = -0.66, R) >droYak2.chr2L 479461 89 - 22324452 --------------ACUGACUCAAUUAGAUAGCCCGUUUCCGCUAAGGAUGAGGCAUUUUGUUUGCGG-AAACGGAAAUGAAUGCAUCAUGCUAAACUUAAGUG --------------.((((.....)))).((((((((((((((..((((((...))))))....))))-))))))..((((.....)))))))).......... ( -25.50, z-score = -2.66, R) >droSec1.super_14 477626 103 - 2068291 ACGUUGAUUCCACUUCUGACUCAAUUGGAUAGCUAAUUUCCGCUGAGGAUGAGCCAUUUUGUUUGCGGAAAACGGAAAUGAAUGGAUCAUGCUAAACUUAAAU- ..(((((.((.......)).)))))....((((........))))((.((((.((((.(..(((.((.....)).)))..))))).)))).))..........- ( -21.50, z-score = -0.68, R) >droSim1.chr2L 501296 103 - 22036055 ACGUUAAUUCCACUUCUGACUCAAUUGGAUAGCUAAUUUCCGCUGAGGAUGAGCCAUUUUGUUUGCGGAAAACGGAAAUGAAUGGAUCAUGCUAAACUUAAAU- ..(((...((((.............))))((((........))))((.((((.((((.(..(((.((.....)).)))..))))).)))).)).)))......- ( -20.82, z-score = -0.66, R) >droPer1.super_1 2596547 96 - 10282868 -----GAUGAUUUUUAUAGCUUAAUU-GCUUGCAGAUCUCCAUCGCUUCUUGAAAGUUGAUUGUAUCCCAAA-AGAAGGGAAGAUAUCAGAUGAAAAUAUAUU- -----((.(((((....(((......-)))...)))))))((((((((.....))))((((....((((...-....))))....))))))))..........- ( -17.60, z-score = -0.09, R) >consensus ACGUUAAUUCCACUUCUGACUCAAUUGGAUAGCUAAUUUCCGCUGAGGAUGAGCCAUUUUGUUUGCGGAAAACGGAAAUGAAUGGAUCAUGCUAAACUUAAAU_ .............................((((........))))((.((((.((((((.((((.((.....)).)))))))))).)))).))........... (-13.00 = -12.32 + -0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:00 2011